[Illinois] ECE 416 Impedance Based Sensors I
[Illinois] ECE 416 Lecture 21: Impedance Based Sensors I
-
 1. Impedance Based Biosensors
0
00:00/00:00
1. Impedance Based Biosensors
0
00:00/00:00 -
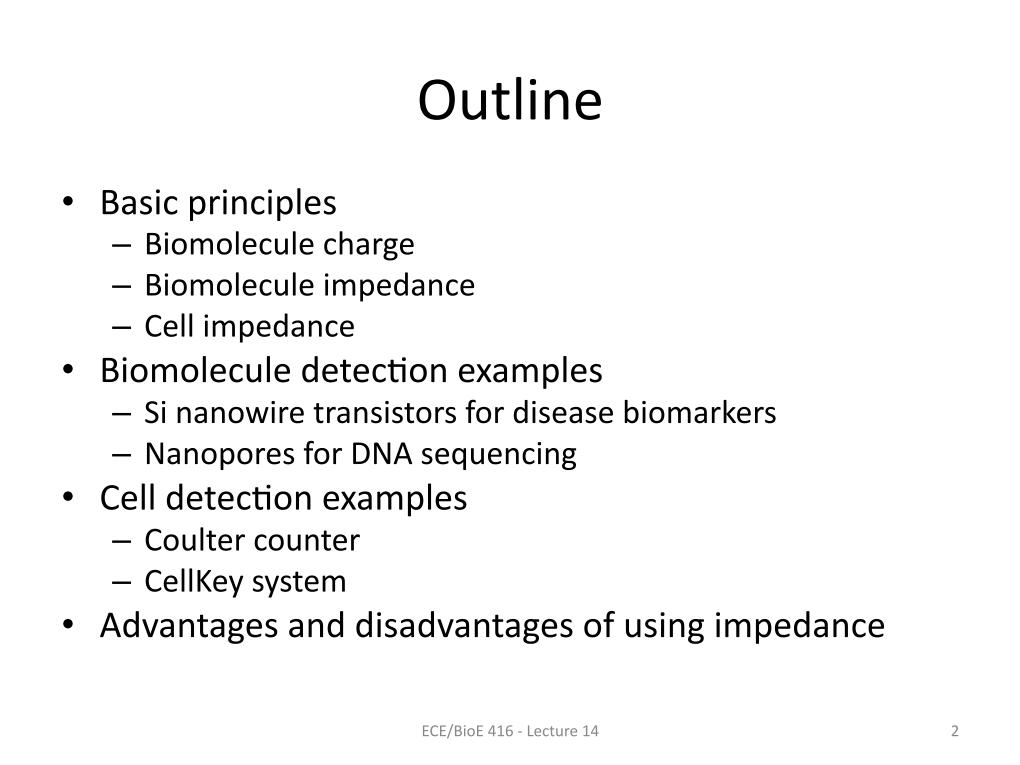 2. Outline
68.177613320999072
00:00/00:00
2. Outline
68.177613320999072
00:00/00:00 -
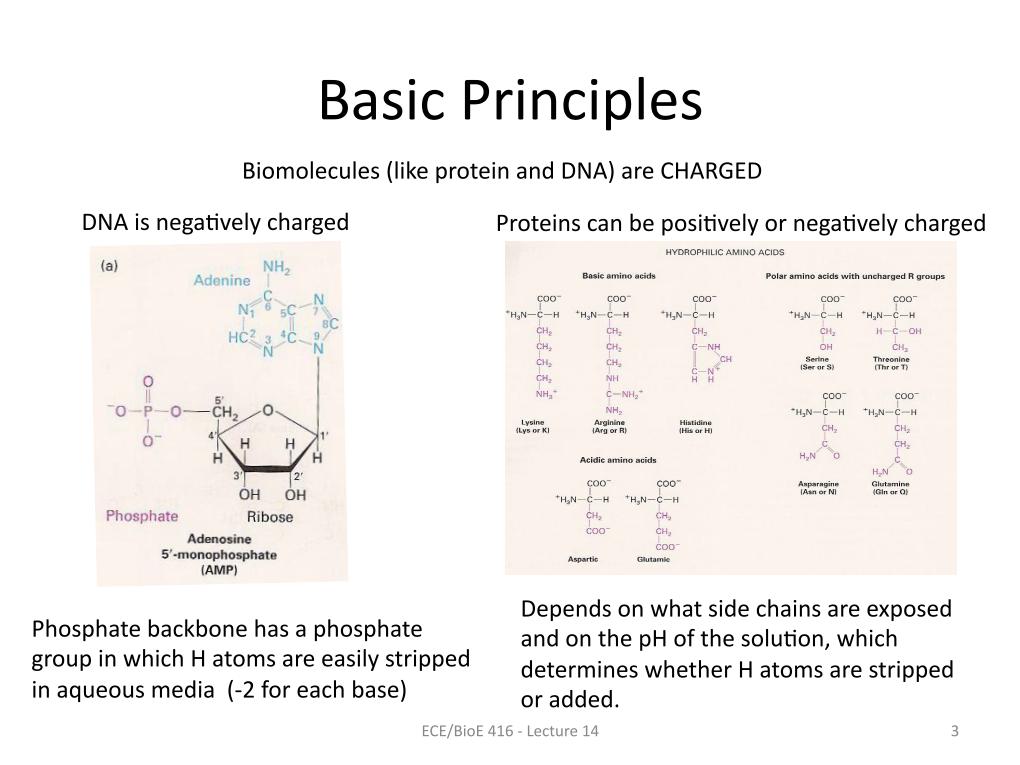 3. Basic Principles
212.35162995594715
00:00/00:00
3. Basic Principles
212.35162995594715
00:00/00:00 -
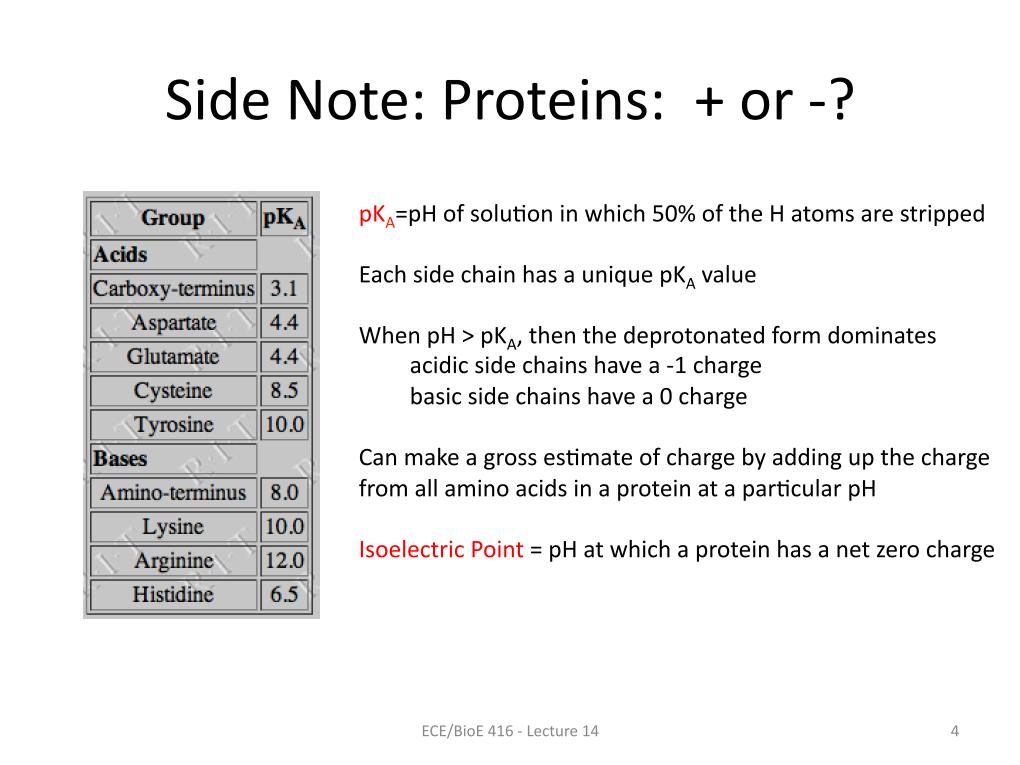 4. Side Note: Proteins: + or -?
431.87225941555346
00:00/00:00
4. Side Note: Proteins: + or -?
431.87225941555346
00:00/00:00 -
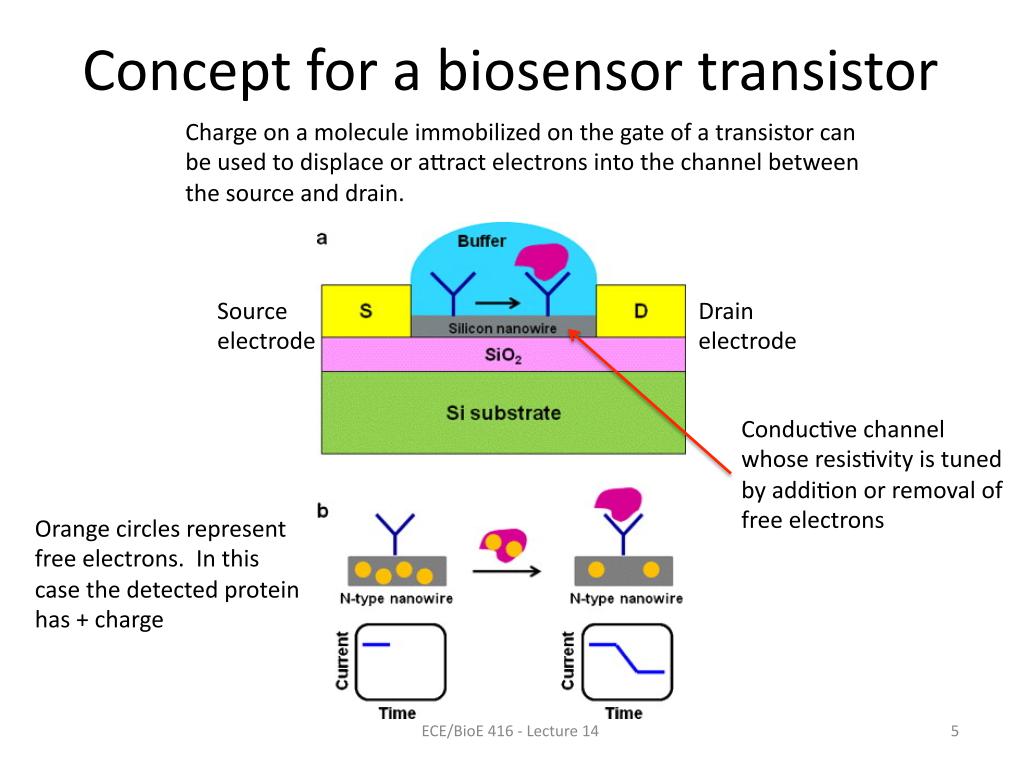 5. Concept for a Biosensor transi…
601.78189548497642
00:00/00:00
5. Concept for a Biosensor transi…
601.78189548497642
00:00/00:00 -
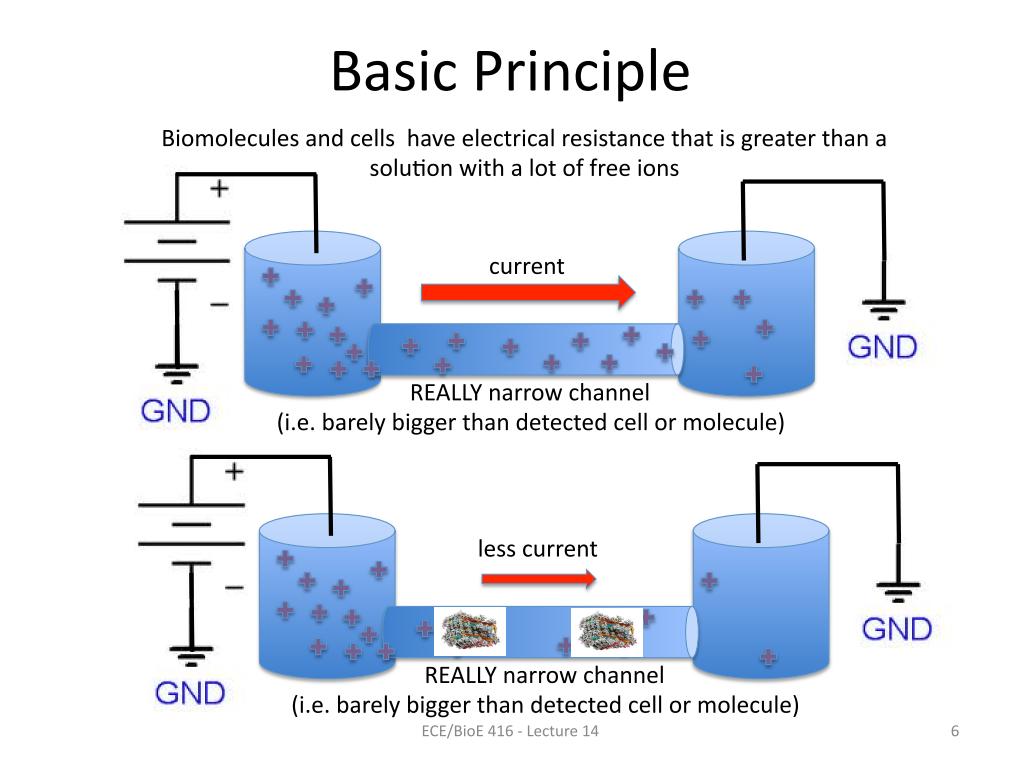 6. Basic Principle
837.58397055557316
00:00/00:00
6. Basic Principle
837.58397055557316
00:00/00:00 -
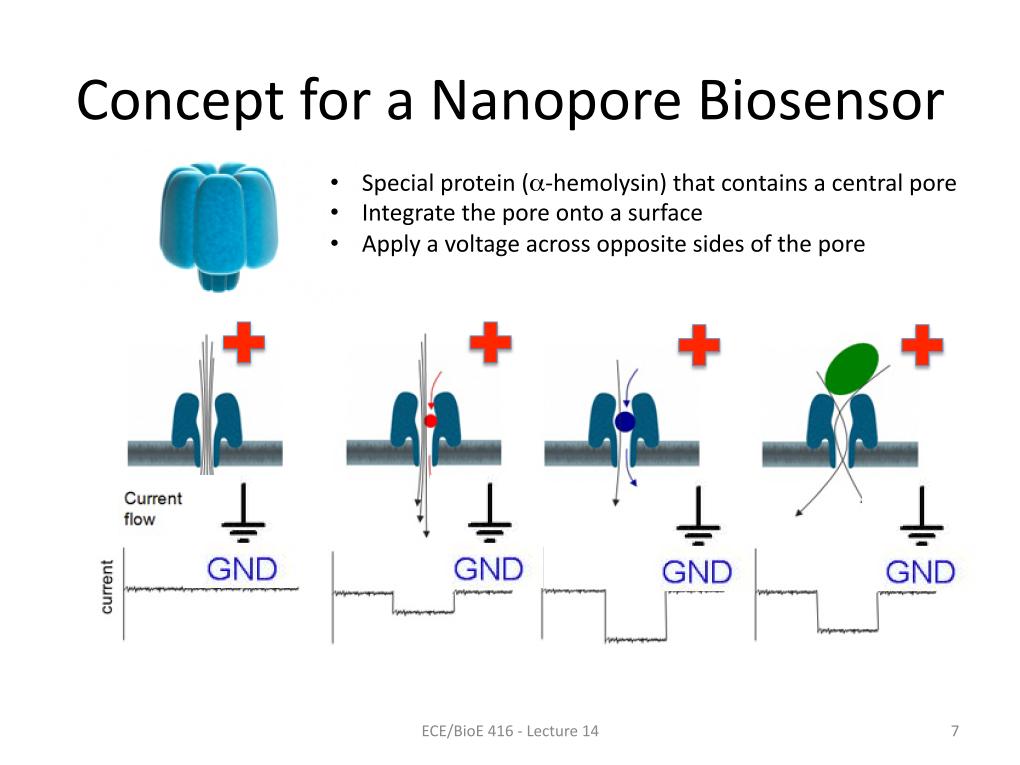 7. Concept for a Nanopore Biosens…
971.77954754576888
00:00/00:00
7. Concept for a Nanopore Biosens…
971.77954754576888
00:00/00:00 -
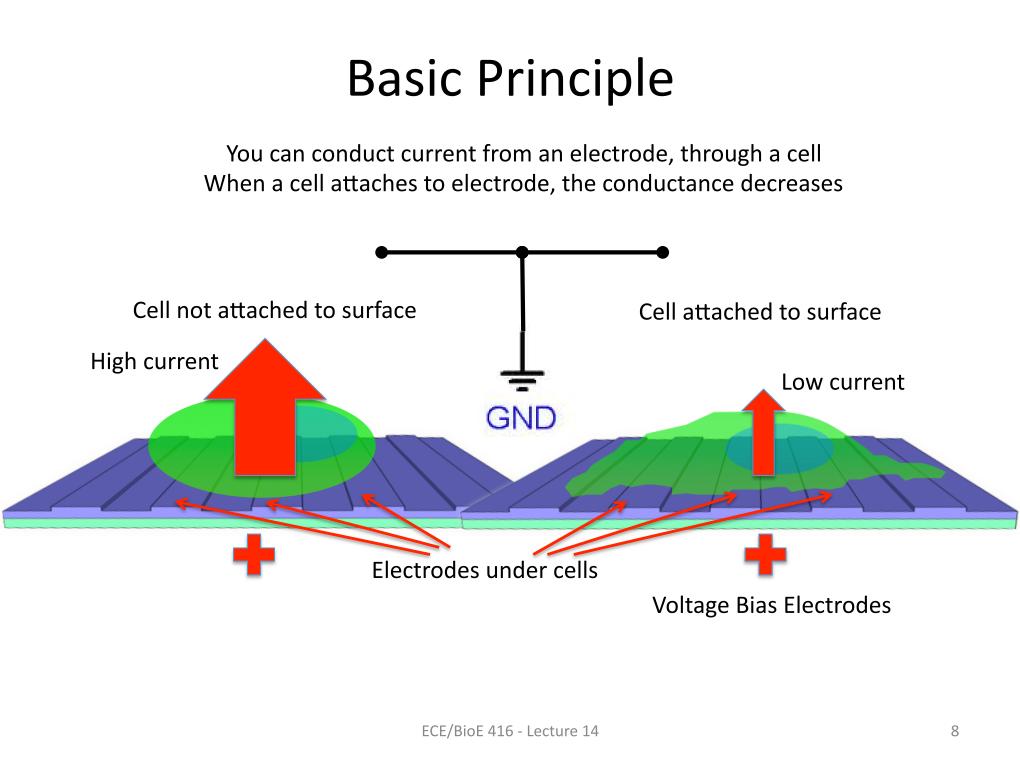 8. Basic Principle
1117.4929085890155
00:00/00:00
8. Basic Principle
1117.4929085890155
00:00/00:00 -
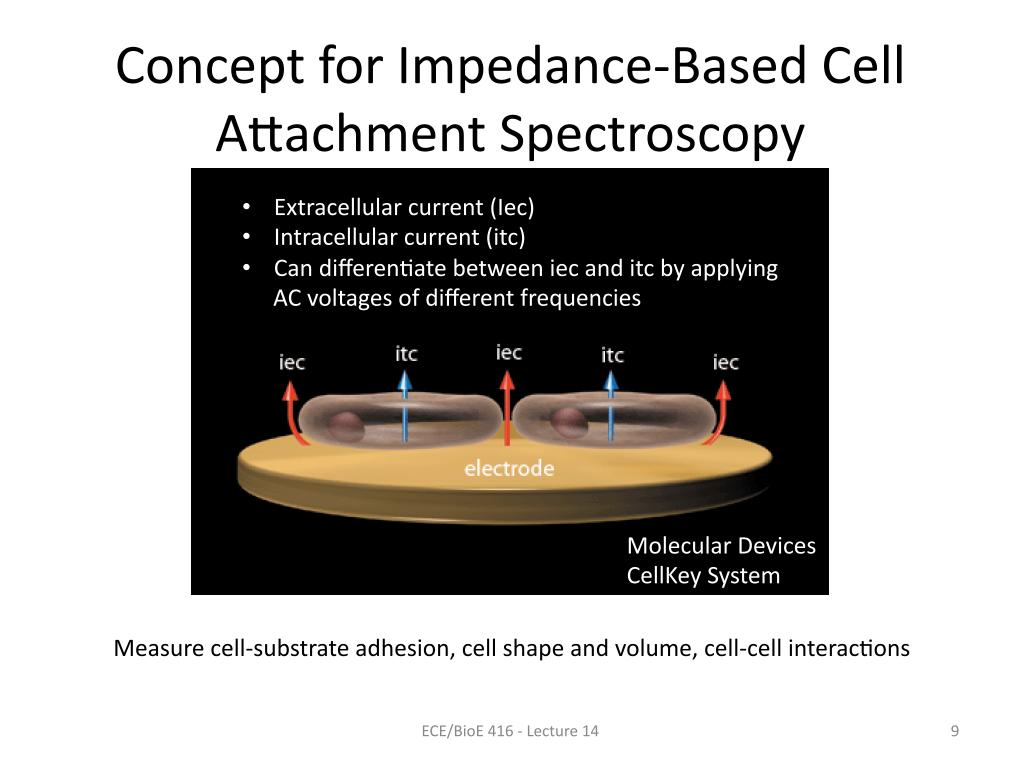 9. Concept for Impedance-Based Ce…
1225.5279373036774
00:00/00:00
9. Concept for Impedance-Based Ce…
1225.5279373036774
00:00/00:00 -
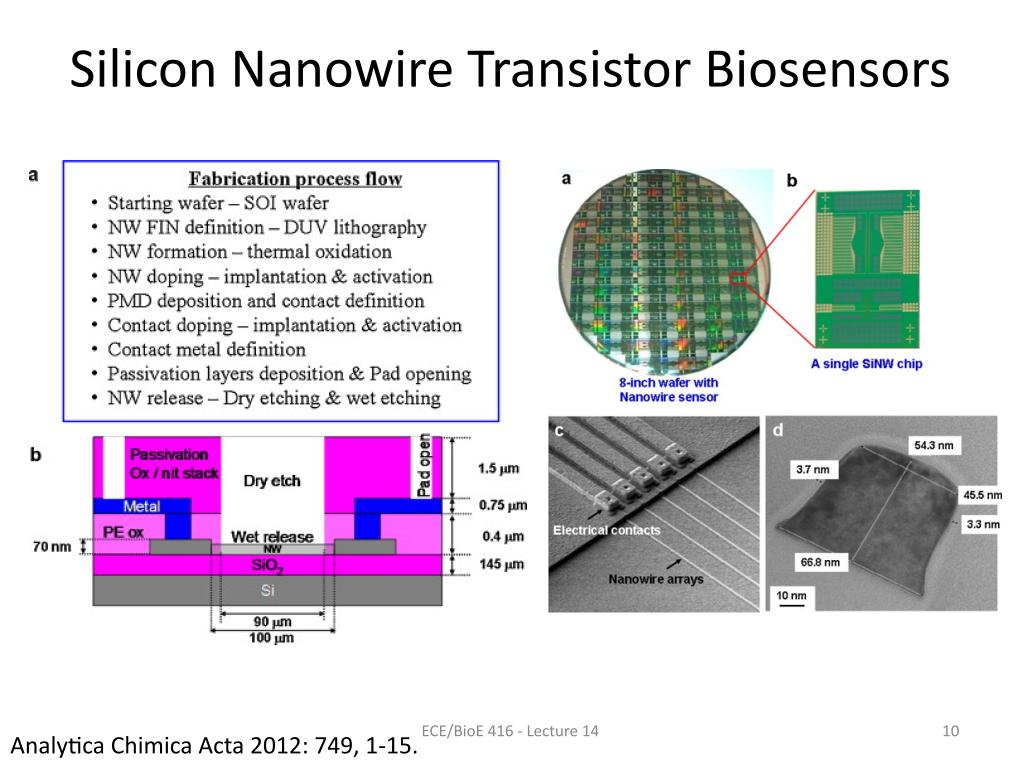 10. Silicon Nanowire Transistor Bi…
1264.9026874385254
00:00/00:00
10. Silicon Nanowire Transistor Bi…
1264.9026874385254
00:00/00:00 -
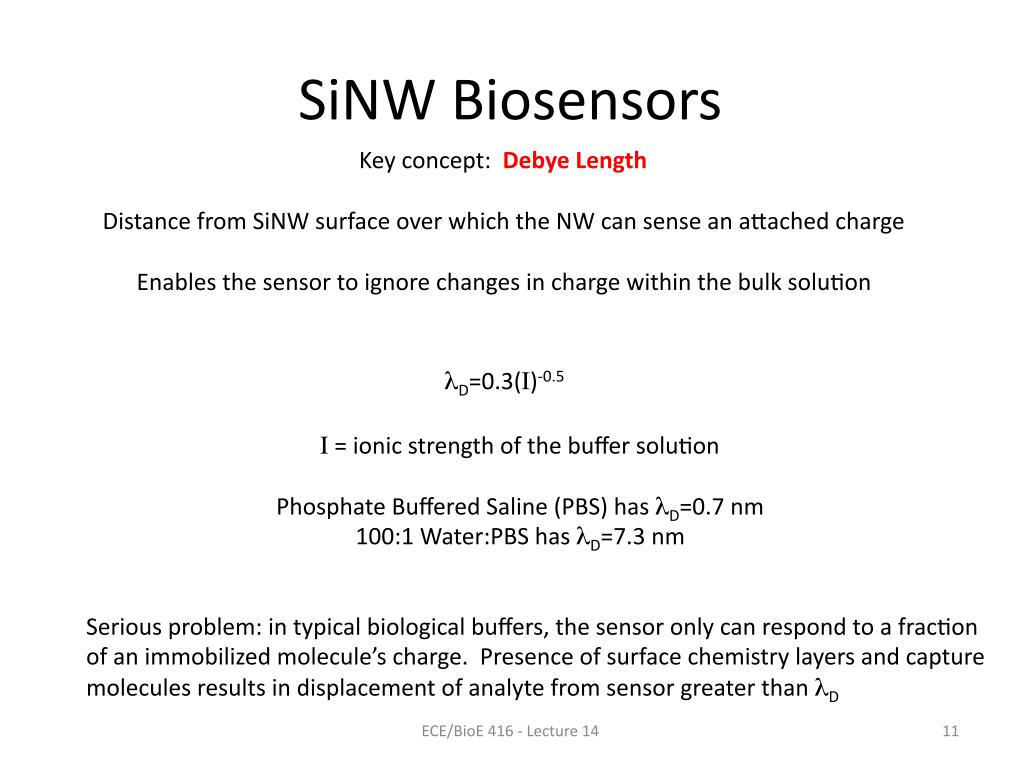 11. SiNW Biosensors Key concept: D…
1469.6335311101943
00:00/00:00
11. SiNW Biosensors Key concept: D…
1469.6335311101943
00:00/00:00 -
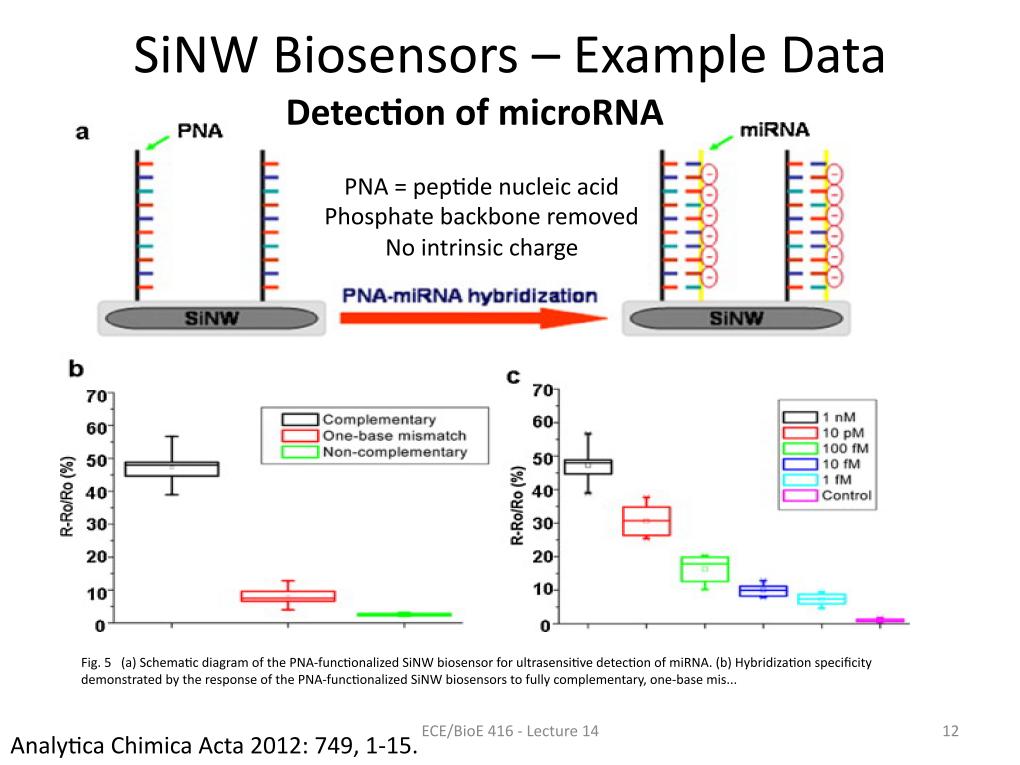 12. SiNW Biosensors
1780.4351302471682
00:00/00:00
12. SiNW Biosensors
1780.4351302471682
00:00/00:00 -
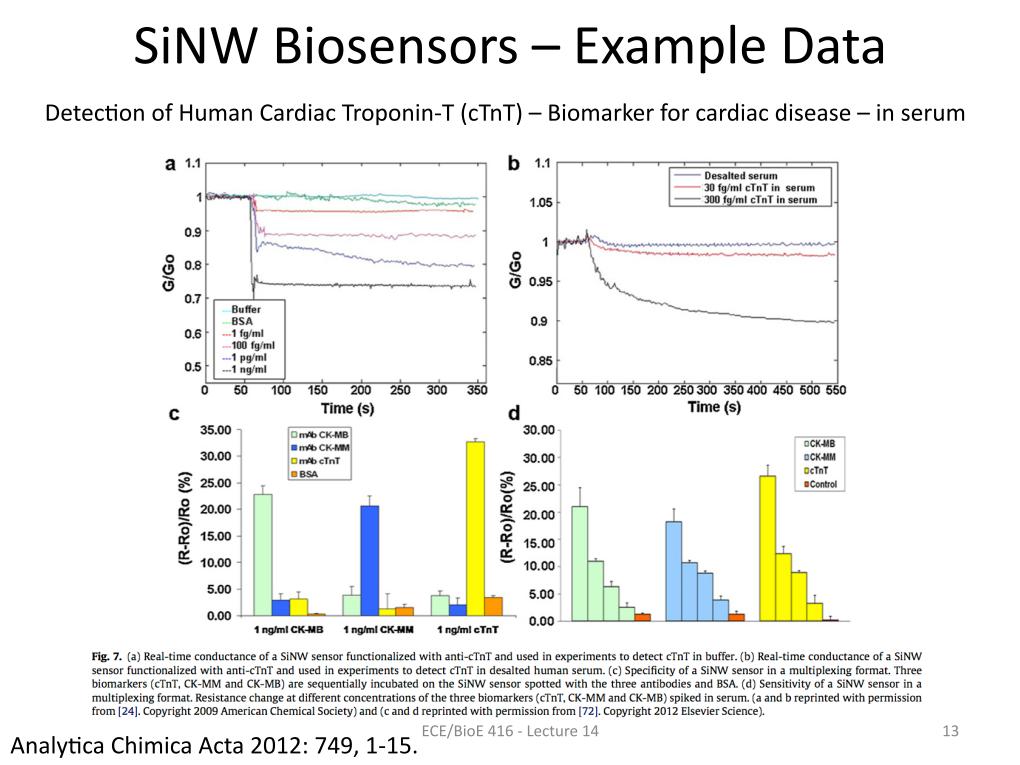 13. SiNW Biosensors
1958.2911444617189
00:00/00:00
13. SiNW Biosensors
1958.2911444617189
00:00/00:00 -
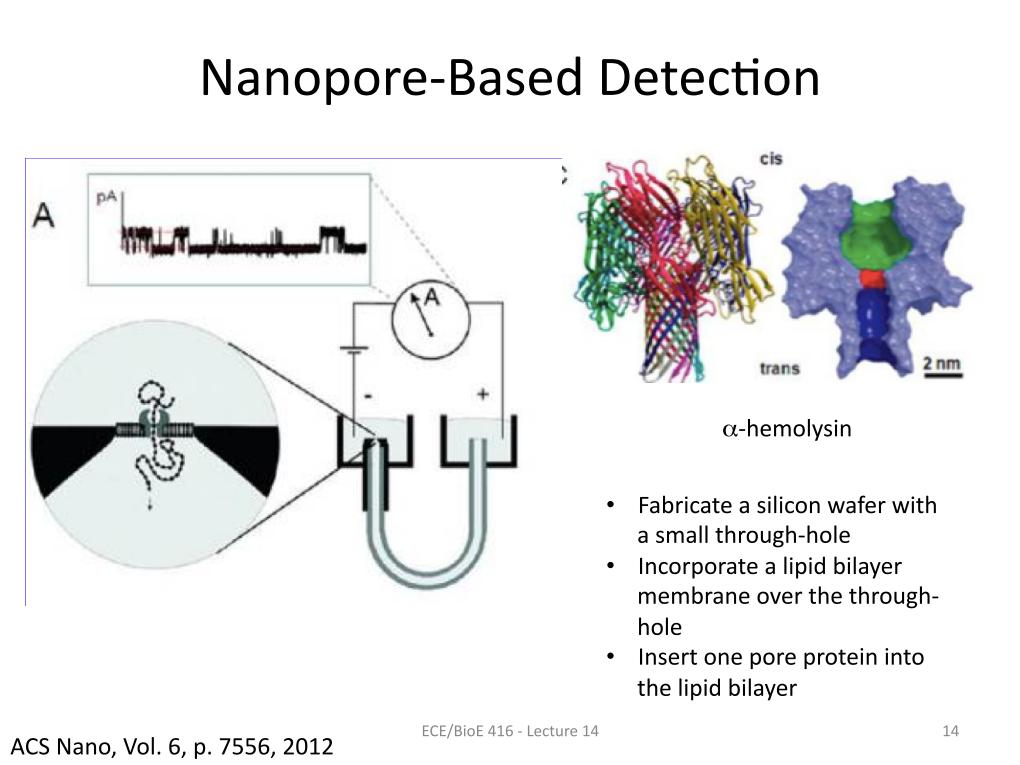 14. Nanopore Based Detection
2085.5224799314656
00:00/00:00
14. Nanopore Based Detection
2085.5224799314656
00:00/00:00 -
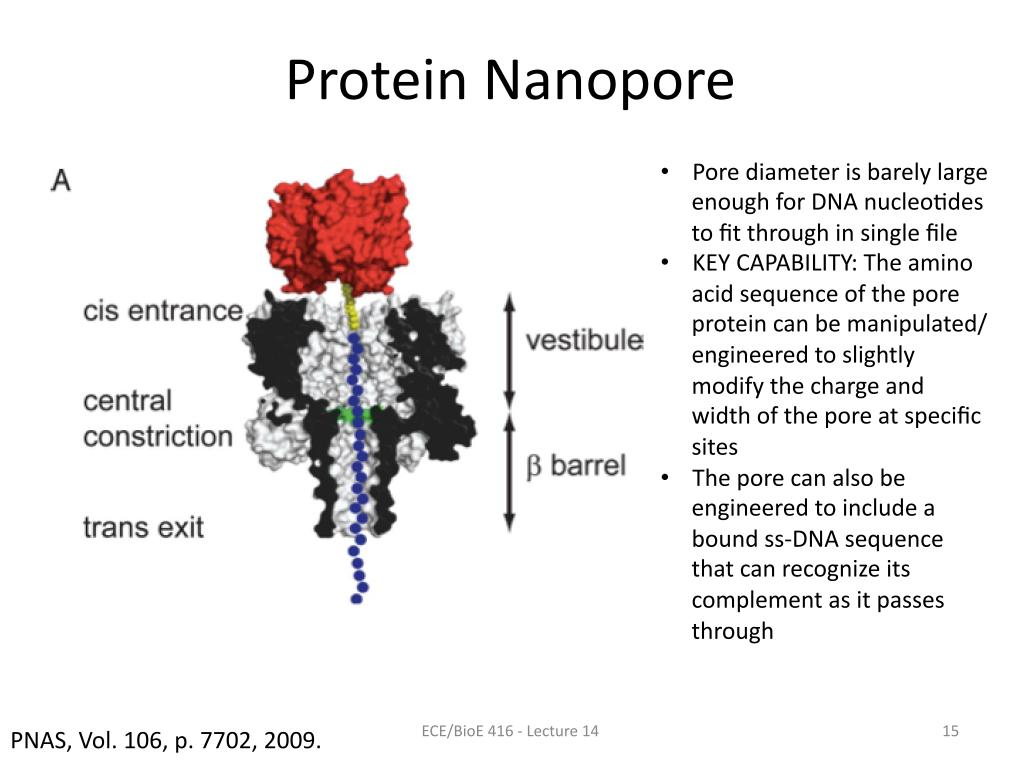 15. Protein Nanopore
2187.4861186026592
00:00/00:00
15. Protein Nanopore
2187.4861186026592
00:00/00:00 -
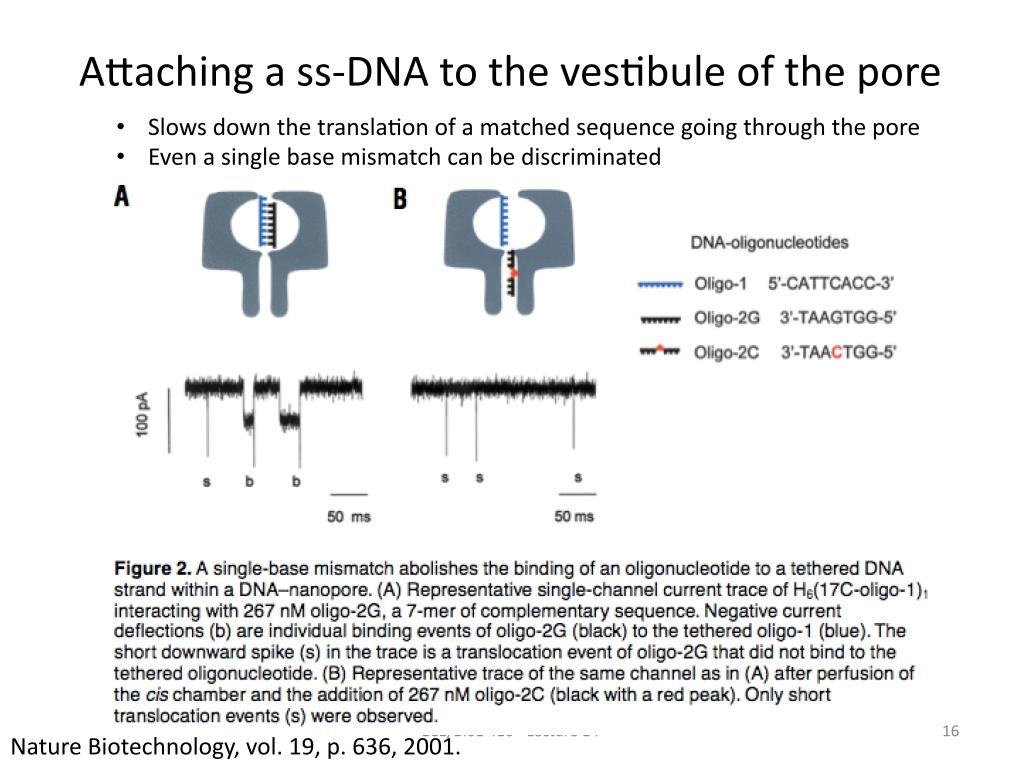 16. Attaching a ss-DNA to the vest…
2290.9676047847192
00:00/00:00
16. Attaching a ss-DNA to the vest…
2290.9676047847192
00:00/00:00 -
 17. Engineered Pore for Differenti…
2425.1631817749153
00:00/00:00
17. Engineered Pore for Differenti…
2425.1631817749153
00:00/00:00 -
 18. Engineered Pore for Differenti…
2529.6268045816541
00:00/00:00
18. Engineered Pore for Differenti…
2529.6268045816541
00:00/00:00 -
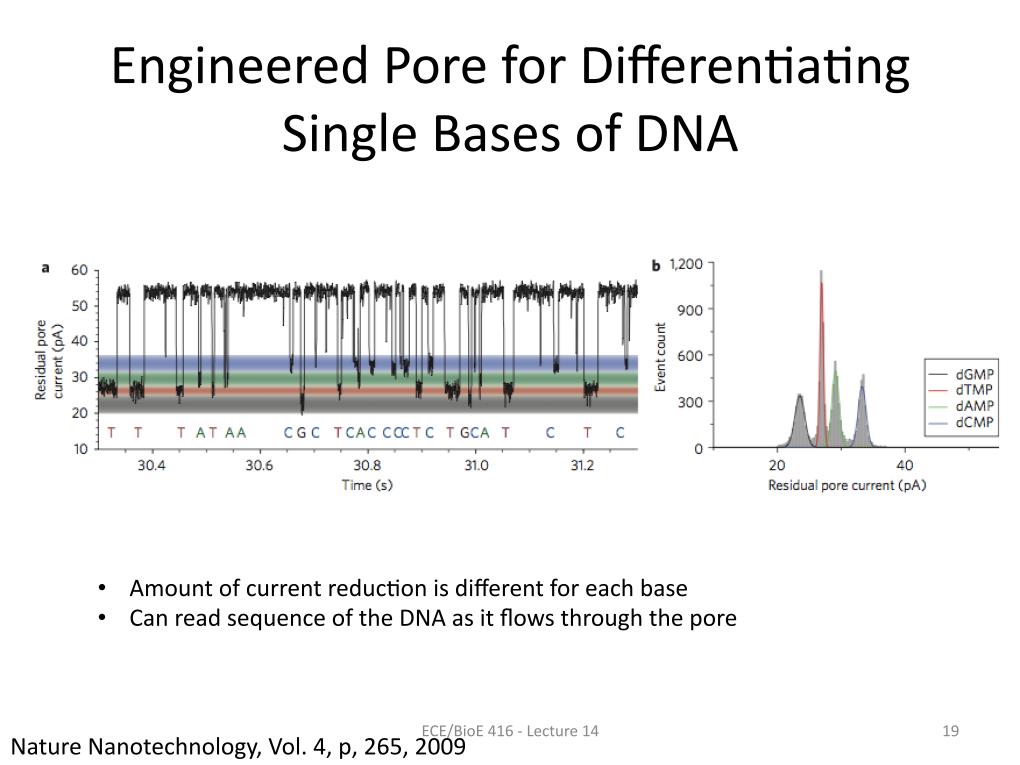 19. Engineered Pore for Differenti…
2555.8766380048864
00:00/00:00
19. Engineered Pore for Differenti…
2555.8766380048864
00:00/00:00 -
 20. Oxford Nanopore Technologies
2613.46555827014
00:00/00:00
20. Oxford Nanopore Technologies
2613.46555827014
00:00/00:00