You must login before you can run this tool.
Category
Published on
Abstract
Hypoxia Simulator
This app demonstrates the evaluation of the new hypotheses on the frequency and duration of phenotypic changes in cancer cells under the influence of hypoxic conditions.
This model and cloud-hosted demo are part of a collaboration between Dr. Macklin's lab and Dr. Gilkes's lab, through of a project granted by Jayne Koskinas Ted Giovanis (JKTG) Foundation for Health and Policy and the Breast Cancer Research Foundation (BCRF). It is also part of the education and outreach for the IU Engineered nanoBIO Node and the NCI-funded cancer systems biology grant U01CA232137. The model is built using PhysiCell: a C++ framework for multicellular systems biology [1].
Legend:
Exploring this model
You can follow these suggestions to familiarize yourself with the model.
1) Run this model using the default parameters.
2) Change the fraction_rsp to 0.1. Note that most green cells (90%) will have less migration.
3) Return the fraction_rsp to 0.5 and change bias_green_rsp to 1.0. Note that all green cells will have high migration, due 100% of green cells have a high bias migration.
4) Keep the parameters defined in (3) and change the hypoxia_pers_time to 300 min. Thus, the green cells do not leave the hypoxic region, but produce a radial growth of the tumor.
5) Now, you can enjoy yourself changing other parameters and creating new responses. ![]()
Modeling description
The model developed and based on a new hypoxia target mapping system that was designed to report a fluorescent switch in low oxygen conditions. Normoxic cells express a red fluorescent protein (DsRed), and after exposure to hypoxia conditions, hypoxia-inducible factors (HIFs) drive a Cre-Loxp mechanism to induce a permanent change to express a green fluorescent protein (GFP) [3]. In this scenario, we show the response of the GFP+ cells to the migratory stimulus from hypoxia, with a focus on understanding the role of phenotypic transience or permanence on cancer invasion
Modeling approach
We used the agent-based model to describe the mechanical and phenotypic behavior of each cell. Associated with the disposition of cells in the microenvironment, we solved the diffusion of oxygen in the medium through BioFVM [2]. The availability of oxygen pressure in the region defines cellular phenotypic changes.
Phenotypic changes
The phenotypic transition from normoxic to hypoxic occurs when the concentration of PO2 () is less than a certain threshold (
). At that moment, a permanent genetic change occurs in the cell, starting the process of GFP expression. This process of protein expression is modeled by
in which and
are protein production rates,
and
are protein degradation rates, and
and
are the genes associate to DsRed and GFP, respectively. Cell death by necrosis is a deterministic event and occurs when oxygen pressure in the cell is less than
.
Proliferation
We modeled cell proliferation using the basic Ki67 cell cycle from PhysiCell [1]. In the model, GFP+ and DsRed+ cells, under the same conditions, have identical proliferation capacity according to biological observations using Ki67 staining.
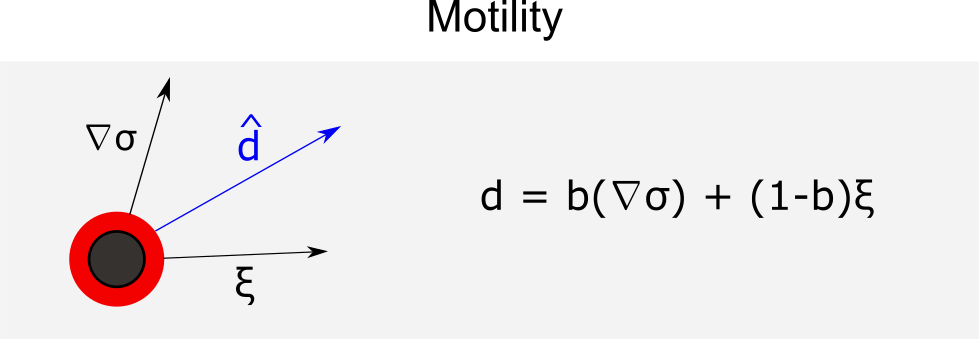
Motility
The cellular motility is characterized in the model by three intrinsic properties:
- Persistence time (
), the average time that the cell maintains its current speed and direction;
- Speed (s), the speed of cellular migration in the absence of other forces;
- Bias (b), a value between 0 and 1 that represents the extent that cell trajectory mimics Brownian motion.
in which represents the random unit vector and
unit vector with the direction of mobility. Thus, the velocity of the cell motility is given by
with the duration of
. To investigate the invasive behavior of GFP+ cells, we define a fraction of that cell (
) that has a different mobility bias (
).
GUI Overview
- Config Basics tab: input parameters common to all models (e.g., domain grid, simulation time, choice/frequency of outputs)
- Microenvironment tab: microenvironment parameters that are model-specific
- User Params tab: user parameters that are model-specific
- Cell Types tab: parameters for cell types that are model-specific
- Out: Plots tab: output display of cells and substrates
- Animate tab: generate an animation of cells
Clicking the 'Run' button will use the specified parameters and start a simulation. When clicked, it creates an "Output" widget that can be clicked/expanded to reveal the progress of the simulation. When the simulation generates output files, they can be visualized in the "Out: Plots" tab. The "# cell frames" will be dynamically updated as those output files are generated by the running simulation. When the "Run" button is clicked, it toggles to a "Cancel" button that will terminate (not pause) the simulation.
Powered by
This software is powered by PhysiCell [1-2], a powerful simulation tool that combines multi-substrate diffusive transport and off-lattice cell models. PhysiCell is BSD-licensed, and available at:
- GitHub releases: https://github.com/MathCancer/PhysiCell/releases
- SourceForge downloads: https://sourceforge.net/projects/physicell/
It is a C++, cross-platform code with minimal software dependencies. It has been tested and deployed in Linux, BSD, OSX, Windows, and other environments, using the standard g++ compiler.
See http://PhysiCell.MathCancer.org.
The Jupyter-based GUI was auto-generated by xml2jupyter [3], a technique to create graphical user interfaces for command-line scientific applications.
References
- Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://dx.doi.org/10.1371/journal.pcbi.1005991
- Ghaffarizadeh A, Friedman SH, Macklin P (2016) BioFVM: an efficient, parallelized diffusive transport solver for 3-D biological simulations. Bioinformatics 32(8):1256-8. https://dx.doi.org/10.1093/bioinformatics/btv730
- Godet, I., Shin, Y. J., Ju, J. A., Ye, I. C., Wang, G., and Gilkes, D. M. (2019). Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis. Nature Communications, 10(1):4862. https://doi.org/10.1038/s41467-019-12412-1
Cite this work
Researchers should cite this work as follows: