You must login before you can run this tool.
Category
Published on
Abstract
Introduction: multicellular system driven by chemical communication
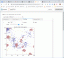
This app demonstrates the complex dynamics that can emerge from (diffusive) chemical communications between multiple cell types. Here, "invader cells" (red) grow in an environment where "suppliers" (yellow) release a resource (σ) that fuels invader birth, and whose absence causes starvation and death in the invaders. Invaders secrete a quorum signal (q) that helps them to aggregate. They also downregulate birth in response to the quorum signal to avoid overcrowding. (Very social cells!) The invader cells release a death signal (ω); live invaders avoid this death signal, as a sign of bad conditions.
But there's more! "Scout" cells (green) migrate along gradients of the quorum signal to find invaders. If they encounter a sufficiently strong quorum signal, they secrete an attack signal (s) that attracts "attackers" (blue). Attackers migrate towards regions of high signal, and activate to release a poison (p) that kills invader cells. But recall that live invaders flee dead invaders. It gets complicated. (It's life.)
Click here to jump straight to the instructions.
This model and cloud-hosted demo are part of a course on computational multicellular systems biology created and taught by Dr. Paul Macklin in the Department of Intelligent Systems Engineering at Indiana University. It is also part of the education and outreach for the IU Engineered nanoBIO Node and the NCI-funded cancer systems biology grant U01CA232137. The models are built using PhysiCell: a C++ framework for multicellular systems biology [1].
Chemical components:
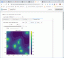
All chemical signals move by chemical diffusion in the simulated environment, using BioFVM [2] to solve the reaction-diffusion equations. A key property is the diffusion length scale. Diffusion (with a diffusion coefficient D) helps spread a signal over large distances, while decay (and uptake) (with coefficient λ) eliminate the signal to slow its spread. These effects compete to determine the characteristic distance L that a chemical signal travels.
That length scale L is given by L = sqrt( D/λ ).
Choosing D and λ can help to tune the chemical communication distance in these models.
Resource (σ):
This is released by supplier agents and consumed by invader agents. Think of it as oxygen or glucose.
The resource is nondimensionalized so that 0 ≤ σ ≤ 1, and we initially assign a length scale of 1000 μm, although cell uptake gives a length scale of 100 μm in densely packed regions.
Quorum (q):
This factor is released by invader cells agents to help coordinate activity.
The resource is nondimensionalized so that 0 ≤ q ≤ 1, and we initially assign a length scale of 100 μm.
Death signal (ω):
This factor is released by dead invader cells agents to signal hostile conditions.
The resource is nondimensionalized so that 0 ≤ ω ≤ 1, and we initially assign a length scale of 200 μm.
Attack signal (s):
This factor is released by scout cells agents to signal that they have detected invaders.
The resource is nondimensionalized so that 0 ≤ s ≤ 1, and we initially assign a length scale of 500 μm.
Poison (p):
Poison is released by attacker cells if they detect a strong attack signal (s).
The resource is nondimensionalized so that 0 < p ≤ 1, and we initially assign a short length scale of 50 μm to reduce collatoral damage to favorable cells. (This is not currently modeled, but it would be a fun twist!)
Cell agents:
We use agent-based models to explore the system. Each cell is modeled as a software agent with an independent state, and its own rules to change its behavior based on local environmental conditions. We use PhysiCell as our modeling platform [1].
Key features in the agent-based models:
Birth and death:
Cells can proliferate (divide into two daughter cells of half size), with division rates regulated by cell rules. In our model, cells continuously work to grow towards a "target" mature volume.
Secretion and uptake:
Cells can secrete chemical factors, or they can remove them (i.e., consume or uptake). This can contribute to gradients in chemical factors, and it's a key method of communication between cells. Moreover, cells can "sample" the chemical state of their surrounding environment. Secreting is sending a signal. Uptaking and sampling is receiving a signal.
Biased migration:
Motile cells perform biased migration. After traveling for Tpersistence time (the mean persistence time), they choose a new migration direction dmigrate. Based on the environmental conditions, the cell chooses a directional bias (intended direction) dbias and a random unit vector r. The motile direction dmigrate is then determined by according to:
dmigrate = b dbias + (1-b) r.
Here, b is a bias parameter (between 0 and 1) that determines how strongly biased migration is towards dbias. If b =1, then migration is deterministic towards dbias. If b = 0, then migration is completely random (Brownian).
Once the migration direction is chosen, it is normalized, and multiplied by the cell's migration speed.
Mechanics:
Cell agents can stick to one another within a prescribed interaction distance (some multiple of their radius), and they can exert a pushing force on neighbors. PhysiCell [1] uses potential functions to implement these simple mechanics. Notably, PhysiCell is an off-lattice model, meaning that cells can have variable sizes, and can move freely with grid artifacts.
They aren't required to move some prescribed number of spatial steps. They also can divide without checking for an open neighbor "site". Instead, they can divide and push their neighbors out of the way.
Invader cells (red)
Invader cells divide at a rate that increases with available resource (σ), and decreases with the quorum signal (q) to avoid overcrowding:
b = bmax ⋅ σ ⋅ (1 - q)
Invader cell death increases when resources (σ) are depleted or in the presence of poison (p). We model this as:
d = dmax ⋅ p ⋅ (1 - σ)
Invader cells release quorum signal (q) when alive, and release the death signal (ω) when dead.
Invader cells use the following as their migration bias direction:
dbias = w ⋅ ∇ q - (1-w) ∇
where w is the weight of the quorum signal, and ∇ is the gradient operator that points towards higher concentrations of a chemical factor.
Think of invader cells as social bacteria.
Scout cells (green)
Scout cells do not divide or die in this simplified model. They choose their migration bias according to
dbias = ∇q.
When q > qthreshold, they secrete signal s.
Think of scouts as macrophages, and s as an inflammatory signal.
Attacker cells (blue);
Attacker cells divide in response to the signal (s) (analogous to T-cell expansion), and die in regions of low signal.
b = bmax ⋅ s
Invader cell death increases when resources (σ) are depleted or in the presence of poison (p). We model this as:
d = dmax ⋅ (1 - s)
They move along towards signals to find targets:
dbias = ∇s.
When s > sthreshold, they secrete poison p.
As a mental model, think of attackers as neutrophils or activated T-cells.
Suppliers (yellow):
Suppliers are inert agents: they don't move, and they can't be moved. They just release resources (). Think of them as cross-sections of blood vessels.
Basic instructions
Modify the parameters in the "config basics" and "user params" tabs. Click the "run" button once ready.
To view the cell plots, click the "cell plots" tab, and slide the bar to advance through simulation frames. Note that as the simulation runs, the "max" field (maximum frame number) will increase, so you can view more simulation frames.
To view the chemical signaling factors, click the "substrate plots" tab, choose a signal from the drop-down menu, and slide through the saved times. Note that as the simulation runs, the "max" field (maximum frame number) will increase, so you can view more simulation frames.
Note that you can download full simulation data for further exploration in your tools of choice.
Note: In most cases, each simulation "frame" should require 2 or 3 seconds. If it's taking much longer to run, the nanoNUB platform (which hosts this tool) is likely under heavy load. You may want to terminate your session and come back later.
Other thoughts
This model was created to show the potential for chemical communication to drive complex dynamics in multicellular systems. It leaves a lot out. In particular, contact-based communication is important in immune-invader dynamics. We'll get back to that in a future module. In teh meantime, I suggest you enjoy this agent-based model of contact- and chemical-based communication in a 3-D PhysiCell model:
Powered by
This software is powered by PhysiCell [1-2], a powerful simulation tool that combines multi-substrate diffusive transport and off-lattice cell models. PhysiCell is BSD-licensed, and available at:
- GitHub releases: https://github.com/MathCancer/PhysiCell/releases
- SourceForge downloads: https://sourceforge.net/projects/physicell/
It is a C++, cross-platform code with minimal software dependencies. It has been tested and deployed in Linux, BSD, OSX, Windows, and other environments, using the standard g++ compiler.
See http://PhysiCell.MathCancer.org.
The Jupyter-based GUI was auto-generated by xml2jupyter [3], a technique to create graphical user interfaces for command-line scientific applications.
Bio
To learn more about our work, please visit MathCancer.org.
Credits
Scientific Staff
Randy Heiland, Research Associate, Intelligent Systems Engineering, Indiana University.
- Lead software architect
Paul Macklin, Ph.D. , Associate Professor, Intelligent Systems Engineering, Indiana University.
- Scientific lead, developed PhysiCell simulation model
The following undergraduate students have contributed to this project:
Jupyter notebook and UI development
- Eric Bower (IU Intelligent Systems Engineering, B.S., 2018-present)
- Daniel Mishler (IU Intelligent Systems Engineering, B.S., 2018-present)
- Tyler Zhang (IU Intelligent Systems Engineering, B.S., 2018)
Sponsored by
- NSF EEC-1720625. Network for Computational Nanotechnology - Engineered nanoBIO Node
- Breast Cancer Research Foundation
- Jayne Koskinas Ted Giovanis Foundation for Health and Policy
- NIH U01CA232137
References
[1] Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://doi.org/10.1371/journal.pcbi.1005991
[2] Ghaffarizadeh A, Friedman SH, Macklin P (2016) BioFVM: an efficient, parallelized diffusive transport solver for 3-D biological simulations. Bioinformatics 32(8):1256-8. https://doi.org/10.1093/bioinformatics/btv730
[3] Heiland R, Mishler D, Zhang T, Bower E, Macklin P (2019, in preparation) Xml2jupyter: Mapping parameters between XML and Jupyter widgets. J Open Source Software.
Cite this work
Researchers should cite this work as follows:
-
This tool is still in preparation for submission. For now, please cite:
Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://doi.org/10.1371/journal.pcbi.1005991