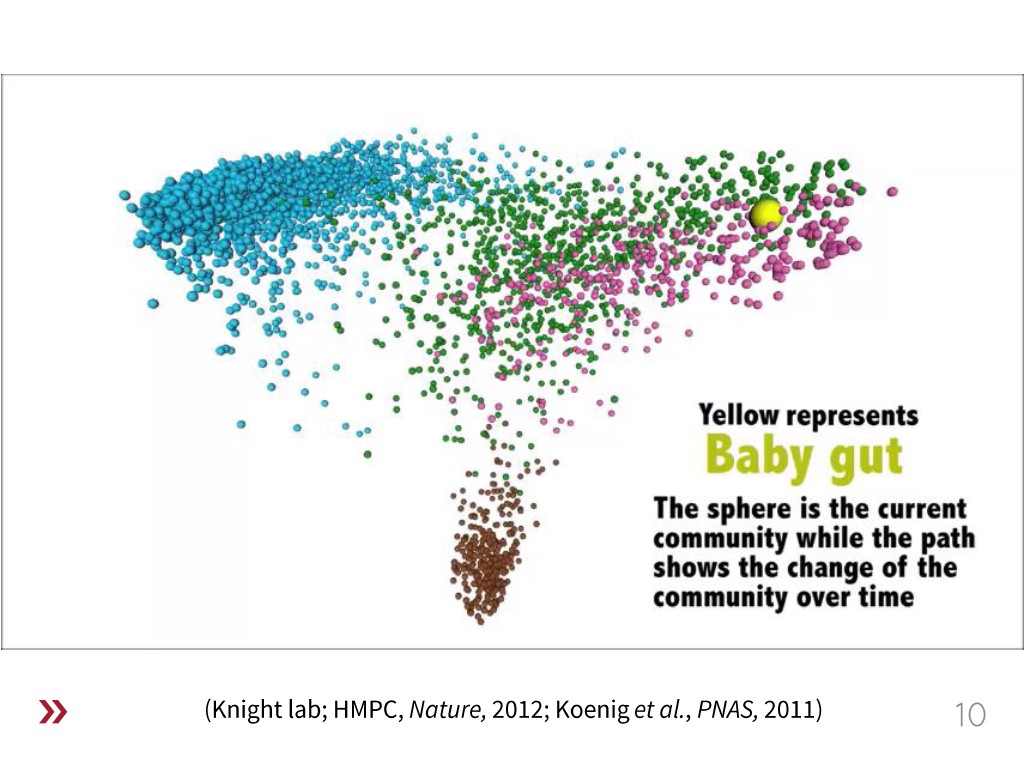
How to Engineer Human Microbiomes?
How to Engineer Human Microbiomes?
-
 1. How to Engineer Human Microbio…
0
00:00/00:00
1. How to Engineer Human Microbio…
0
00:00/00:00 -
 2. Overview
13.146479813146481
00:00/00:00
2. Overview
13.146479813146481
00:00/00:00 -
 3. My Background
31.097764431097765
00:00/00:00
3. My Background
31.097764431097765
00:00/00:00 -
 4. History
72.6393059726393
00:00/00:00
4. History
72.6393059726393
00:00/00:00 -
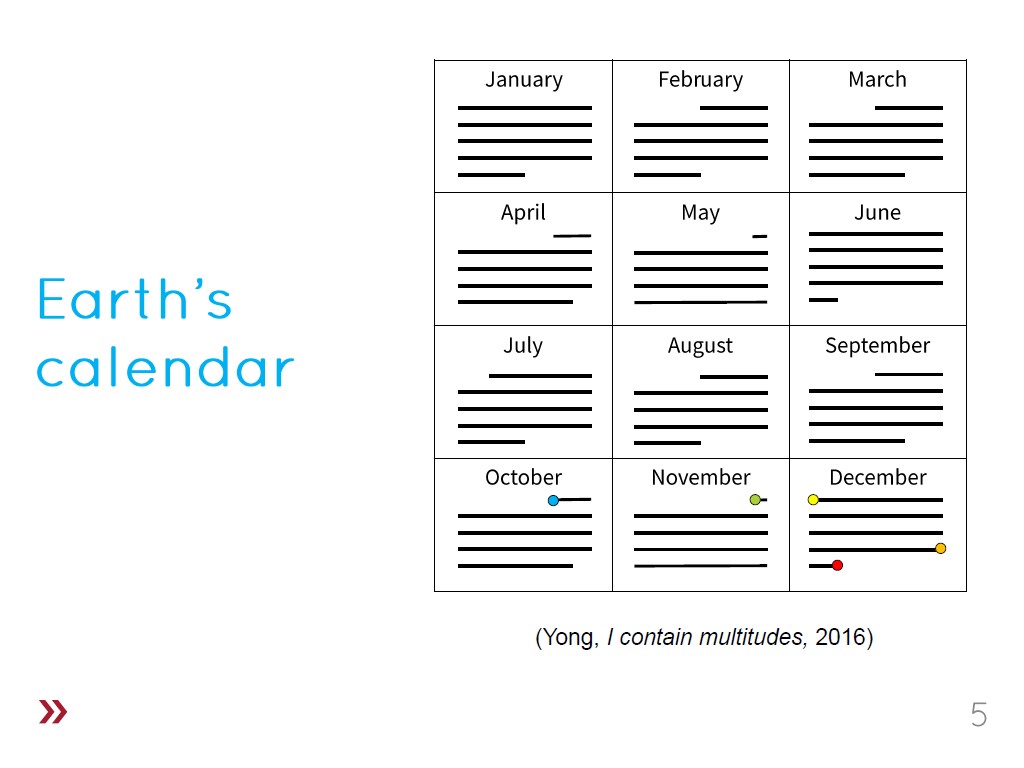 5. Earth's Calendar
76.376376376376385
00:00/00:00
5. Earth's Calendar
76.376376376376385
00:00/00:00 -
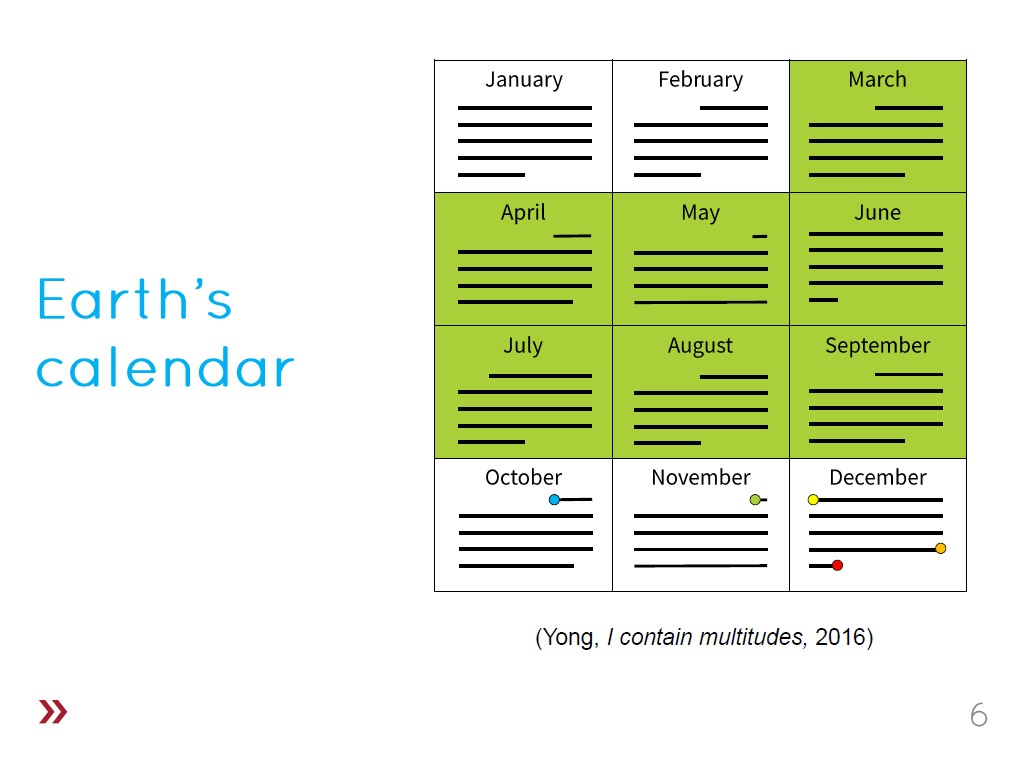 6. Earth's Calendar
132.43243243243245
00:00/00:00
6. Earth's Calendar
132.43243243243245
00:00/00:00 -
 7. Humans
149.01568234901569
00:00/00:00
7. Humans
149.01568234901569
00:00/00:00 -
 8. ~100 trillion bacteria in your…
156.69002335669003
00:00/00:00
8. ~100 trillion bacteria in your…
156.69002335669003
00:00/00:00 -
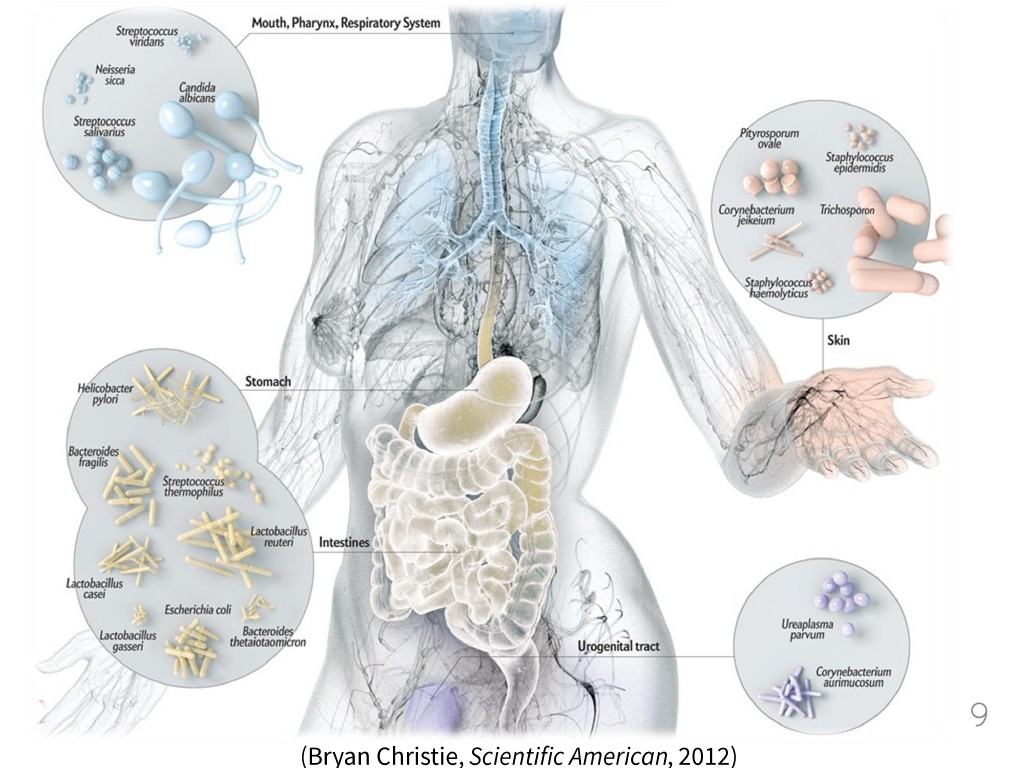 9. Bacteria
171.1378044711378
00:00/00:00
9. Bacteria
171.1378044711378
00:00/00:00 -
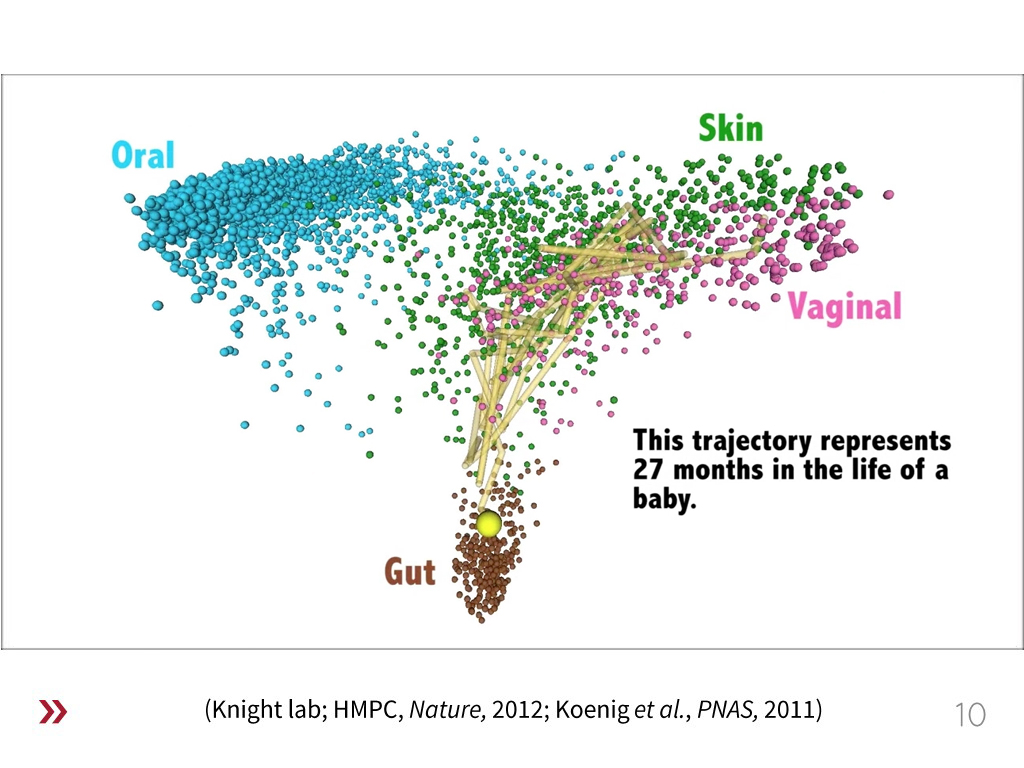 10. Bacteria
202.20220220220222
00:00/00:00
10. Bacteria
202.20220220220222
00:00/00:00 -
 11. Human milk oligosaccharides
277.6443109776443
00:00/00:00
11. Human milk oligosaccharides
277.6443109776443
00:00/00:00 -
 12. Health conditions linked to th…
316.5165165165165
00:00/00:00
12. Health conditions linked to th…
316.5165165165165
00:00/00:00 -
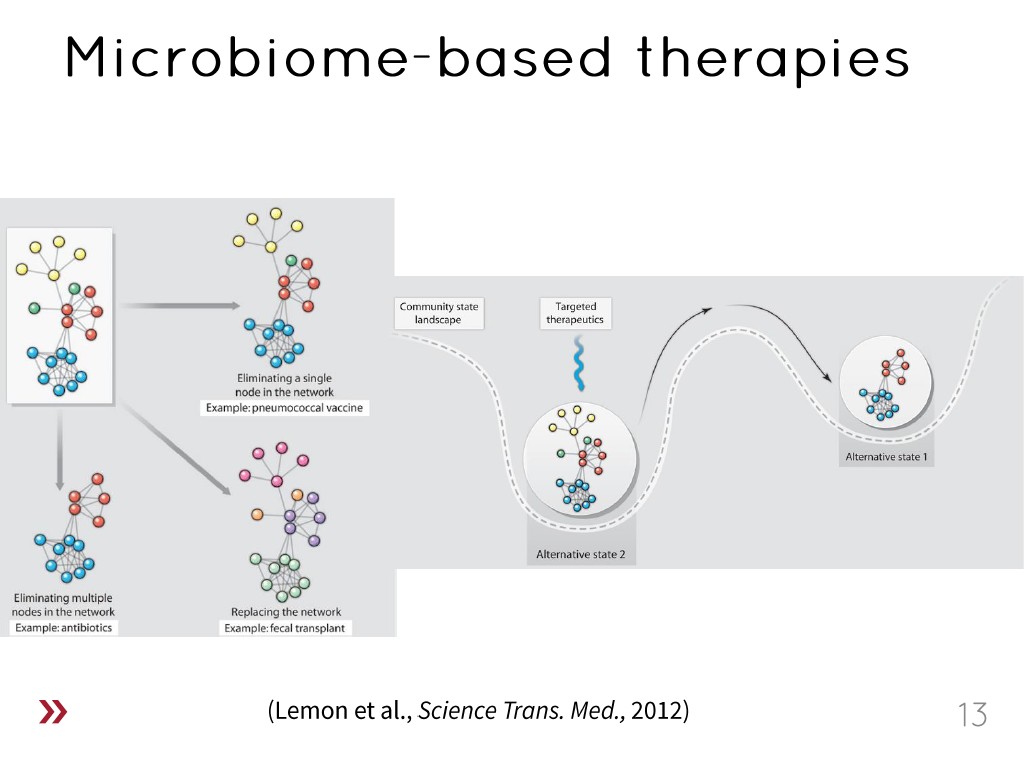 13. Microbiome-based therapies
358.29162495829166
00:00/00:00
13. Microbiome-based therapies
358.29162495829166
00:00/00:00 -
 14. Cause-effect
433.96730063396734
00:00/00:00
14. Cause-effect
433.96730063396734
00:00/00:00 -
 15. Research Overview: Controlled …
463.06306306306305
00:00/00:00
15. Research Overview: Controlled …
463.06306306306305
00:00/00:00 -
 16. Eye drops
475.84250917584251
00:00/00:00
16. Eye drops
475.84250917584251
00:00/00:00 -
 17. Agriculture
598.69869869869876
00:00/00:00
17. Agriculture
598.69869869869876
00:00/00:00 -
 18. Research Overview: Soft Lithog…
677.27727727727734
00:00/00:00
18. Research Overview: Soft Lithog…
677.27727727727734
00:00/00:00 -
 19. Buckling Pneumatic Linear Actu…
685.95261928595266
00:00/00:00
19. Buckling Pneumatic Linear Actu…
685.95261928595266
00:00/00:00 -
 20. Bacterial Networks on Hydropho…
731.86519853186519
00:00/00:00
20. Bacterial Networks on Hydropho…
731.86519853186519
00:00/00:00 -
 21. Diagnostics
804.57123790457126
00:00/00:00
21. Diagnostics
804.57123790457126
00:00/00:00 -
 22. Bacteria are everywhere
817.5175175175176
00:00/00:00
22. Bacteria are everywhere
817.5175175175176
00:00/00:00 -
 23. Pathogens
825.59225892559232
00:00/00:00
23. Pathogens
825.59225892559232
00:00/00:00 -
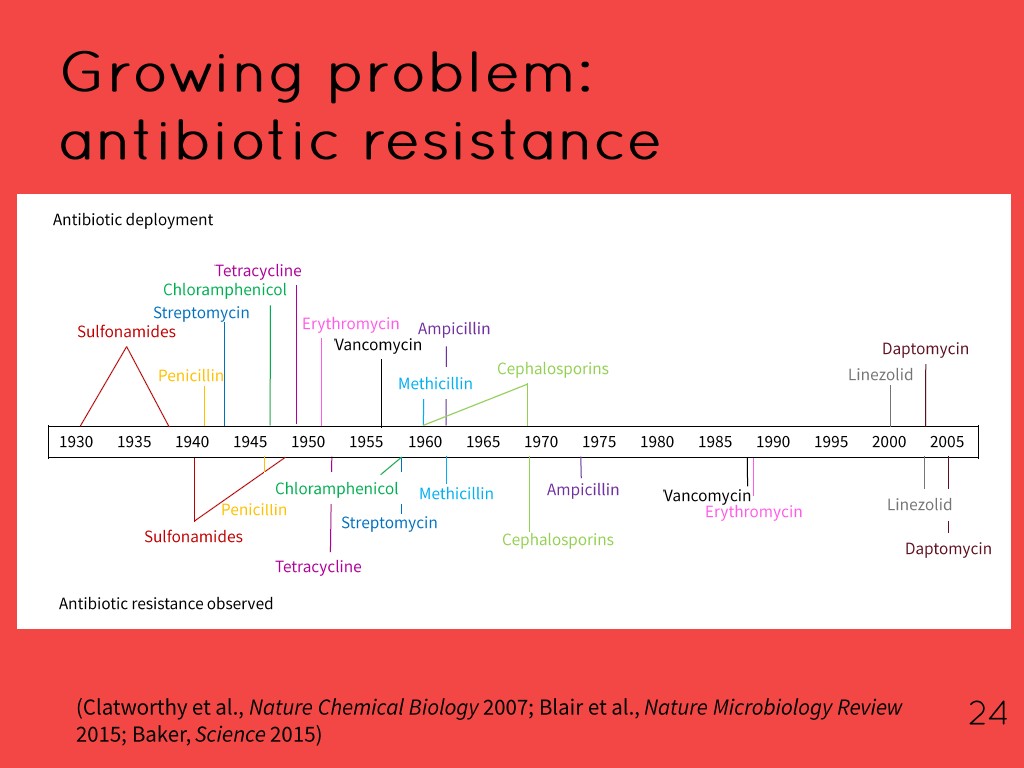 24. Growing problem: antibiotic re…
850.28361695028366
00:00/00:00
24. Growing problem: antibiotic re…
850.28361695028366
00:00/00:00 -
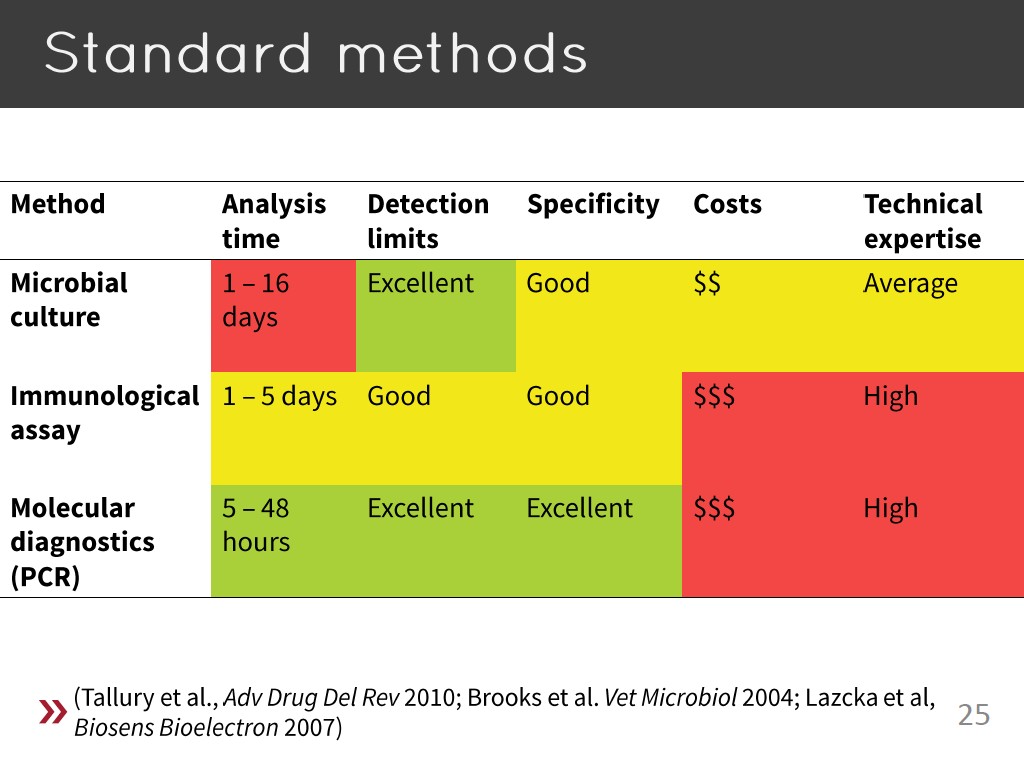 25. Standard Methods
888.42175508842183
00:00/00:00
25. Standard Methods
888.42175508842183
00:00/00:00 -
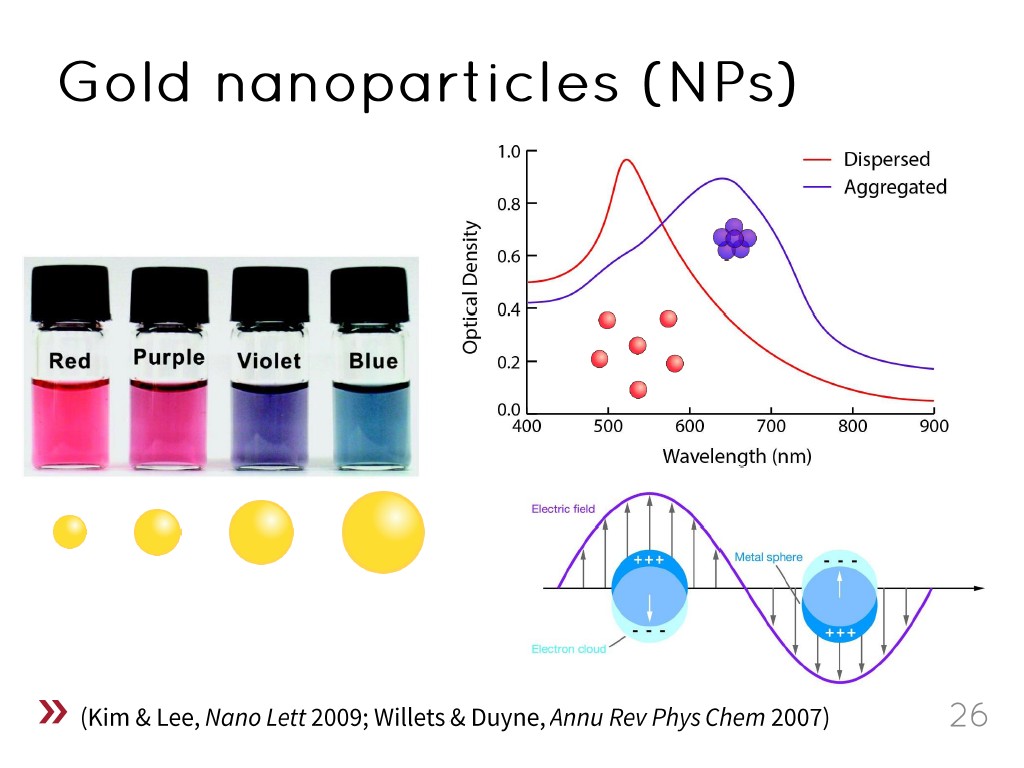 26. Gold nanoparticles (NPs)
941.207874541208
00:00/00:00
26. Gold nanoparticles (NPs)
941.207874541208
00:00/00:00 -
 27. Drawback of existing biosensor…
1037.9045712379045
00:00/00:00
27. Drawback of existing biosensor…
1037.9045712379045
00:00/00:00 -
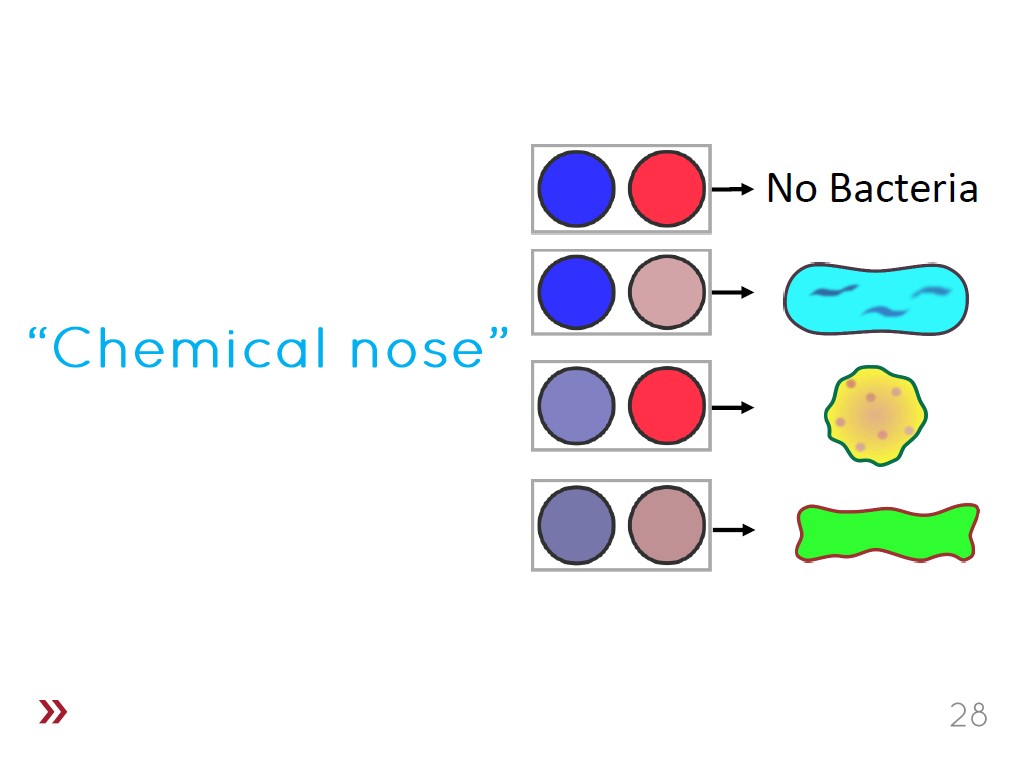 28. Chemical nose
1055.2886219552886
00:00/00:00
28. Chemical nose
1055.2886219552886
00:00/00:00 -
 29. How do we make a Chemical Nose…
1114.8815482148816
00:00/00:00
29. How do we make a Chemical Nose…
1114.8815482148816
00:00/00:00 -
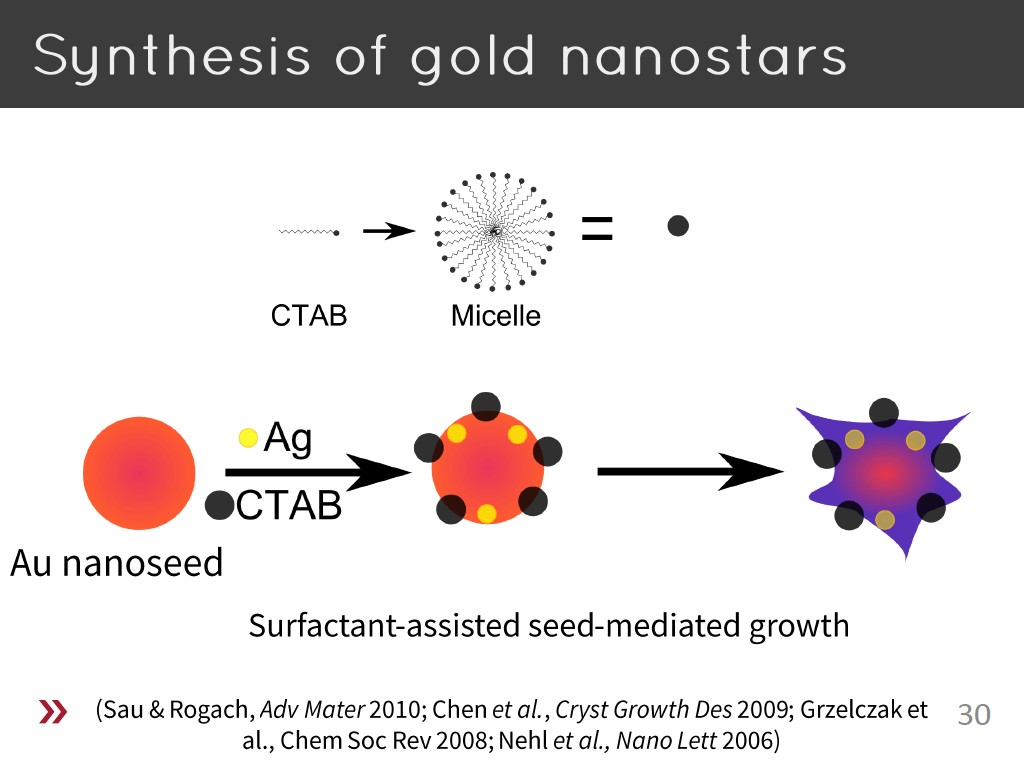 30. Synthesis of gold nanostars
1123.9572906239573
00:00/00:00
30. Synthesis of gold nanostars
1123.9572906239573
00:00/00:00 -
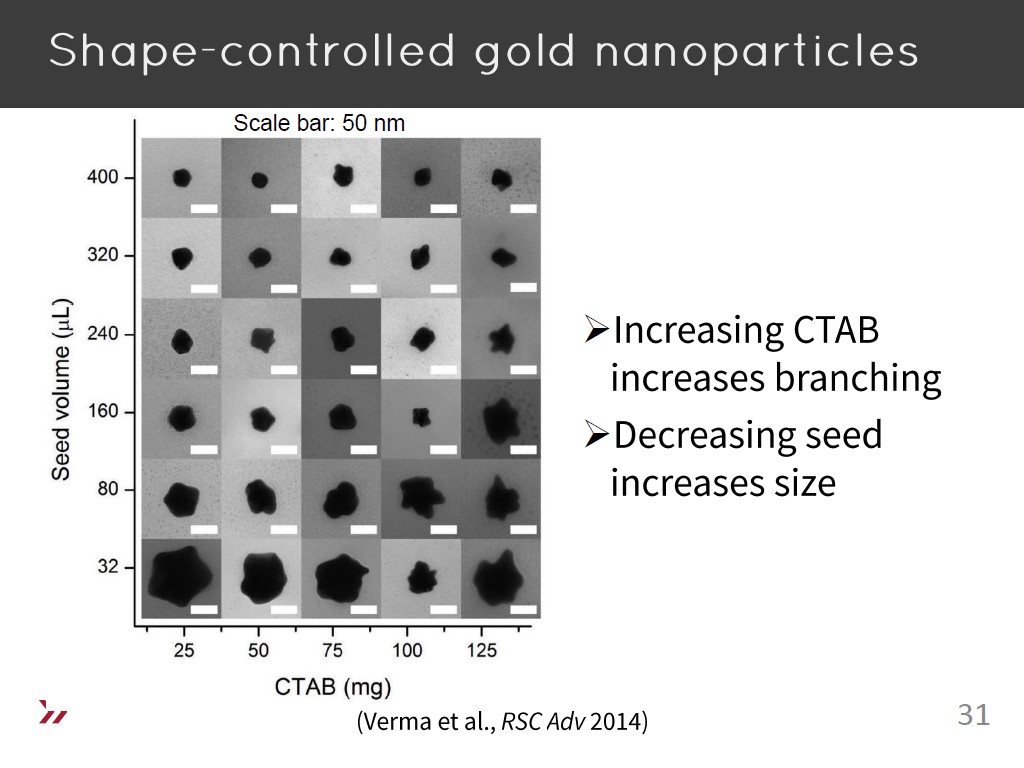 31. Shape-controlled gold nanopart…
1164.0306973640306
00:00/00:00
31. Shape-controlled gold nanopart…
1164.0306973640306
00:00/00:00 -
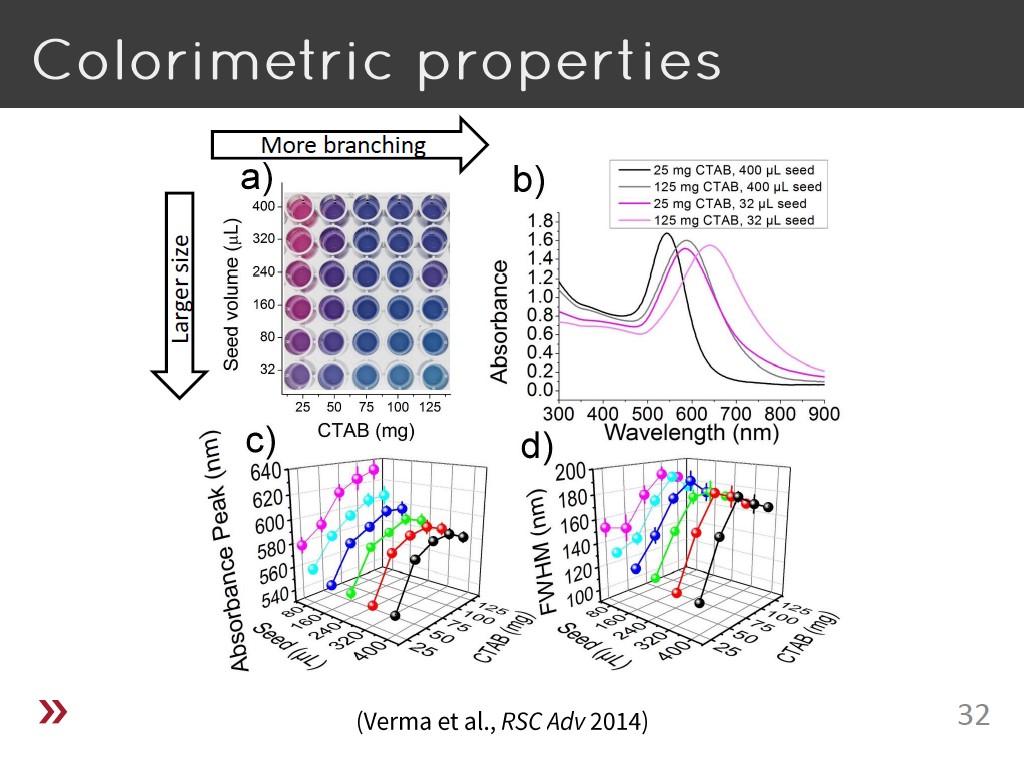 32. Colorimetric properties
1189.7564230897565
00:00/00:00
32. Colorimetric properties
1189.7564230897565
00:00/00:00 -
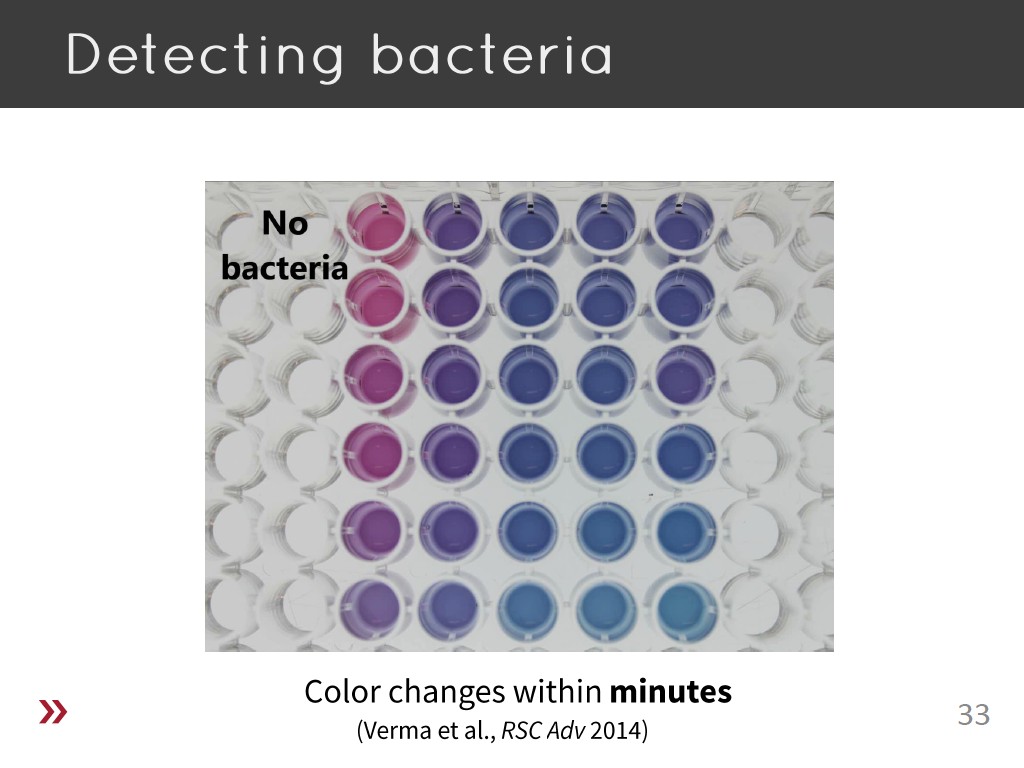 33. Detecting bacteria
1234.2008675342008
00:00/00:00
33. Detecting bacteria
1234.2008675342008
00:00/00:00 -
 34. Demonstrations of Chemical Nos…
1262.9963296629965
00:00/00:00
34. Demonstrations of Chemical Nos…
1262.9963296629965
00:00/00:00 -
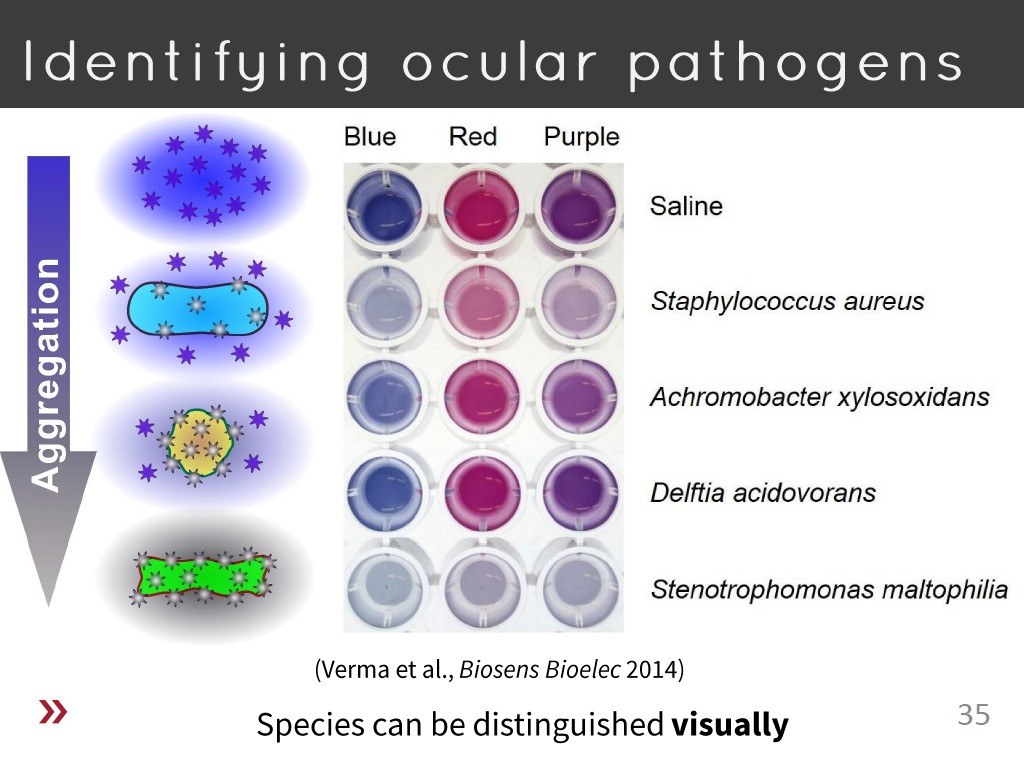 35. Identifying ocular pathogens
1267.0003336670004
00:00/00:00
35. Identifying ocular pathogens
1267.0003336670004
00:00/00:00 -
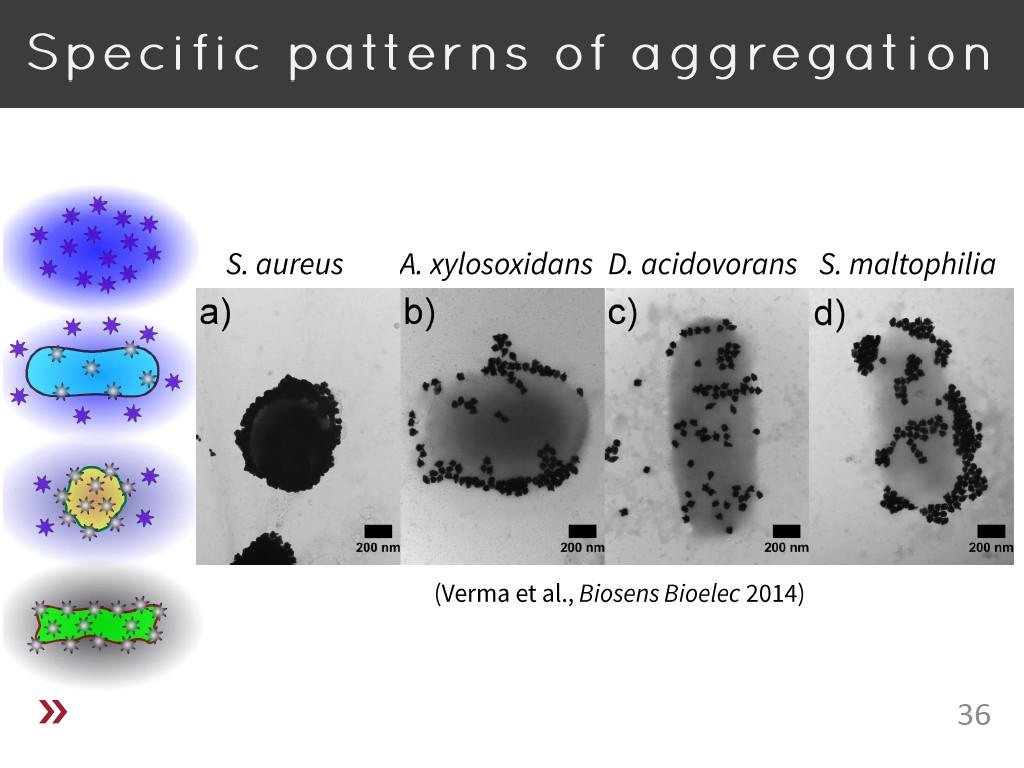 36. Specific patterns of aggregati…
1304.371037704371
00:00/00:00
36. Specific patterns of aggregati…
1304.371037704371
00:00/00:00 -
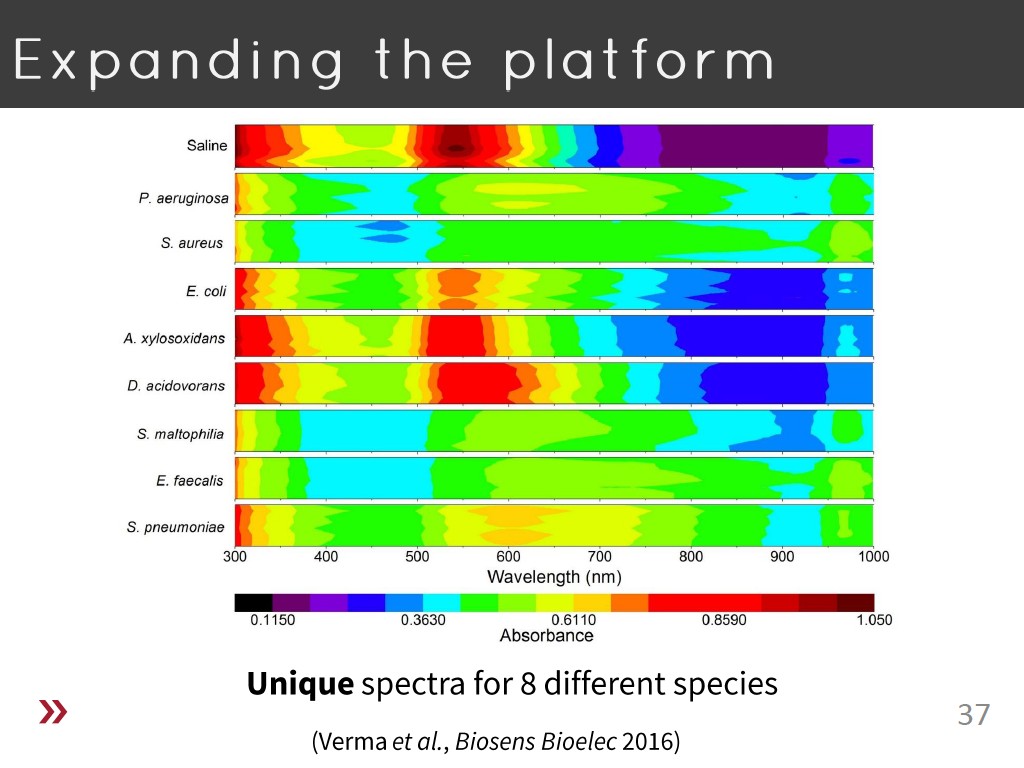 37. Expanding the platform
1339.4394394394394
00:00/00:00
37. Expanding the platform
1339.4394394394394
00:00/00:00 -
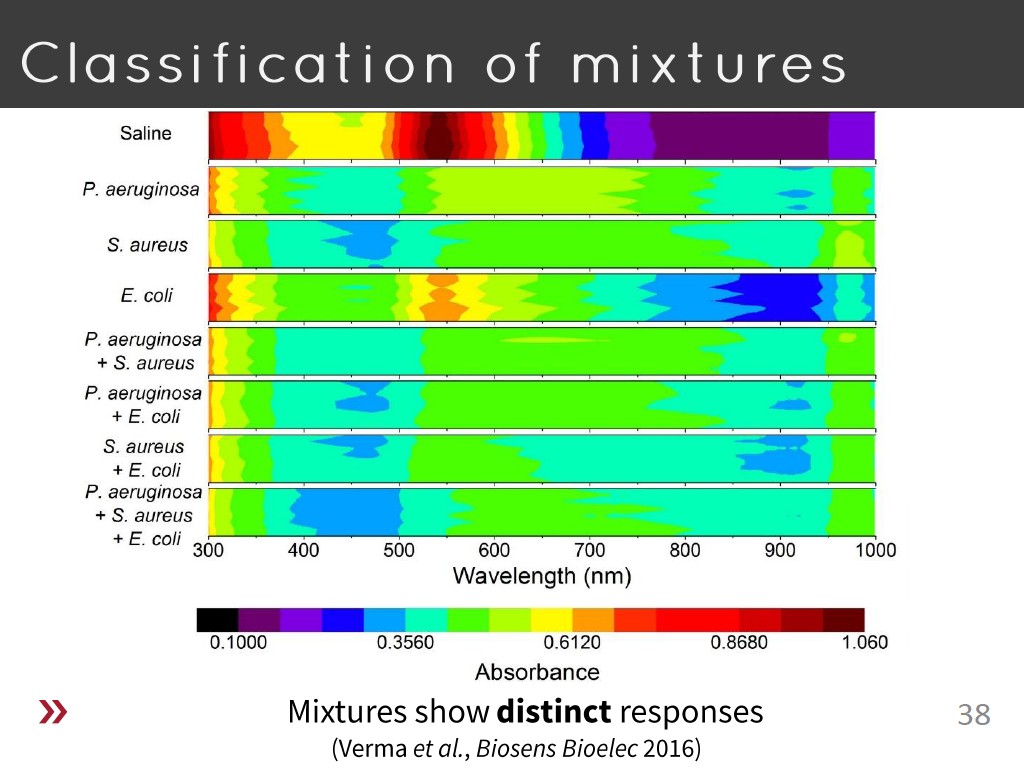 38. Classification of mixtures
1376.6766766766768
00:00/00:00
38. Classification of mixtures
1376.6766766766768
00:00/00:00 -
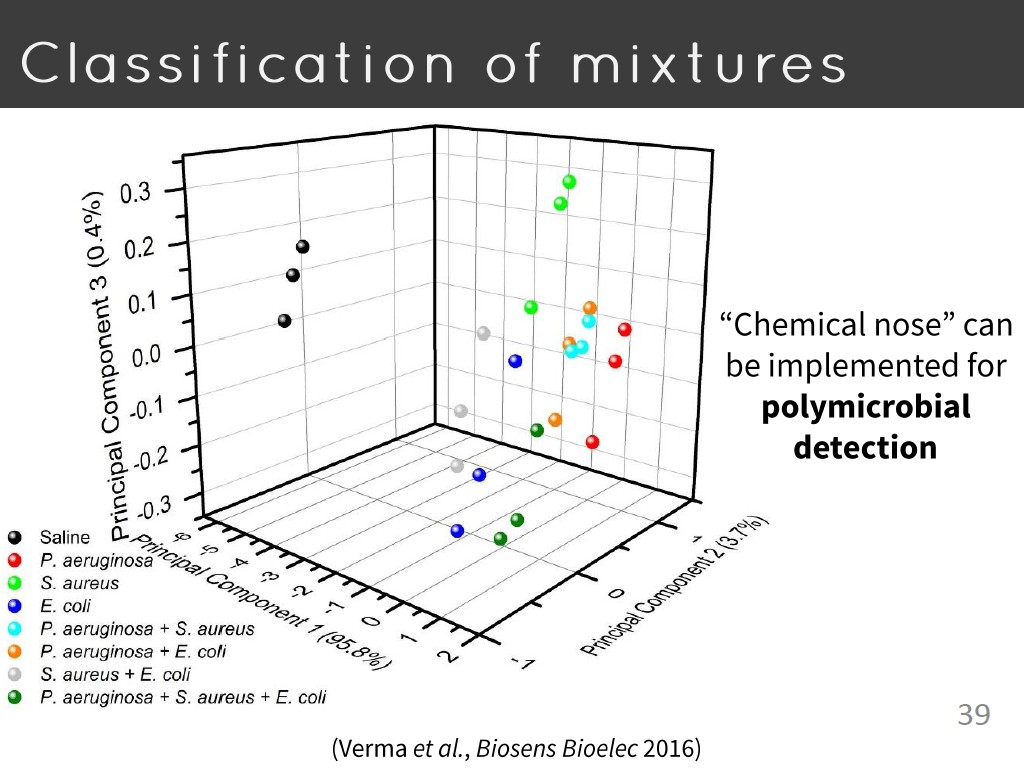 39. Classification of mixtures
1405.6723390056725
00:00/00:00
39. Classification of mixtures
1405.6723390056725
00:00/00:00 -
 40. Exploit shapes for versatility
1429.4961628294961
00:00/00:00
40. Exploit shapes for versatility
1429.4961628294961
00:00/00:00 -
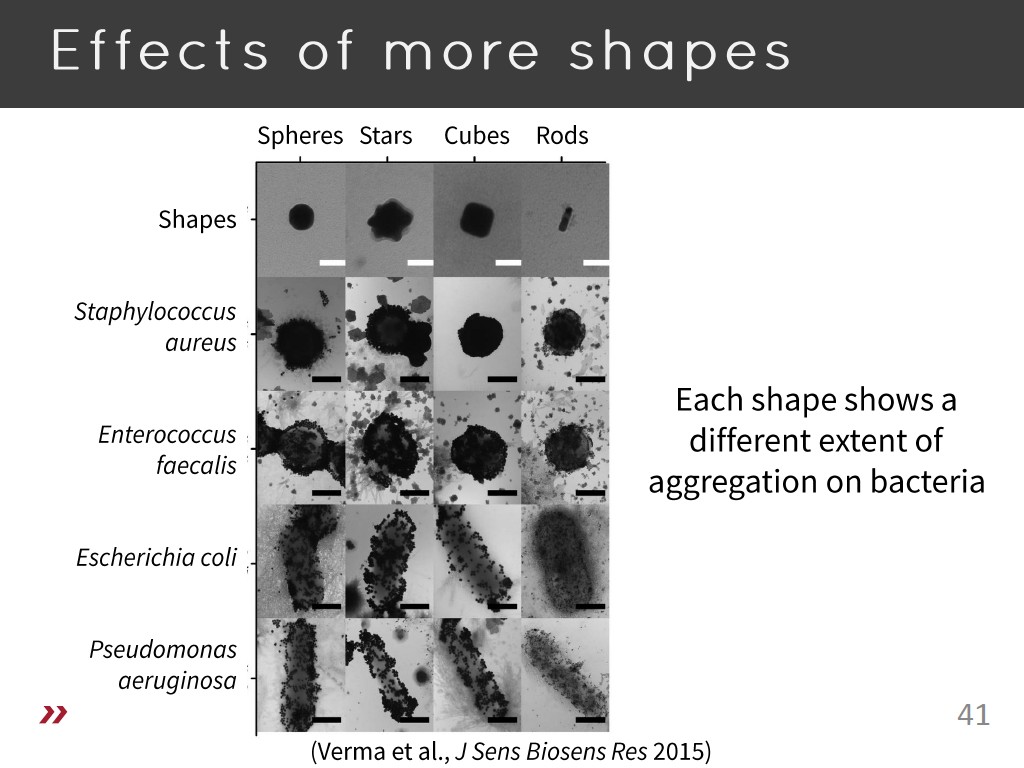 41. Effects of more shapes
1437.303970637304
00:00/00:00
41. Effects of more shapes
1437.303970637304
00:00/00:00 -
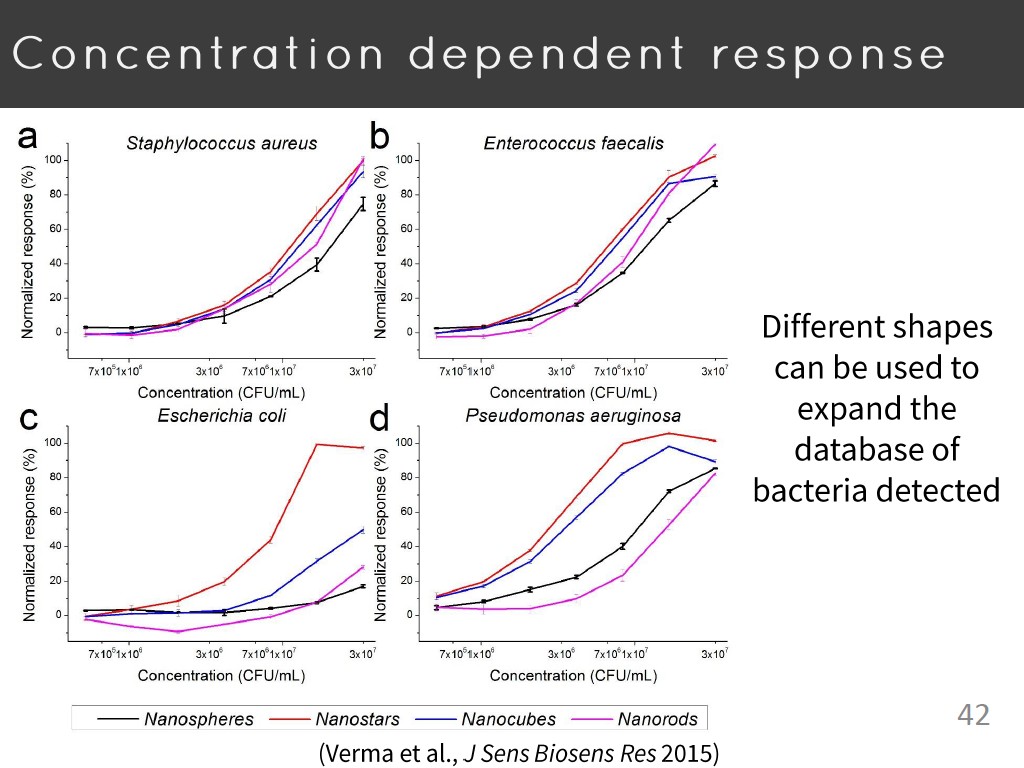 42. Concentration dependent respon…
1472.238905572239
00:00/00:00
42. Concentration dependent respon…
1472.238905572239
00:00/00:00 -
 43. Why does it work?
1521.1211211211212
00:00/00:00
43. Why does it work?
1521.1211211211212
00:00/00:00 -
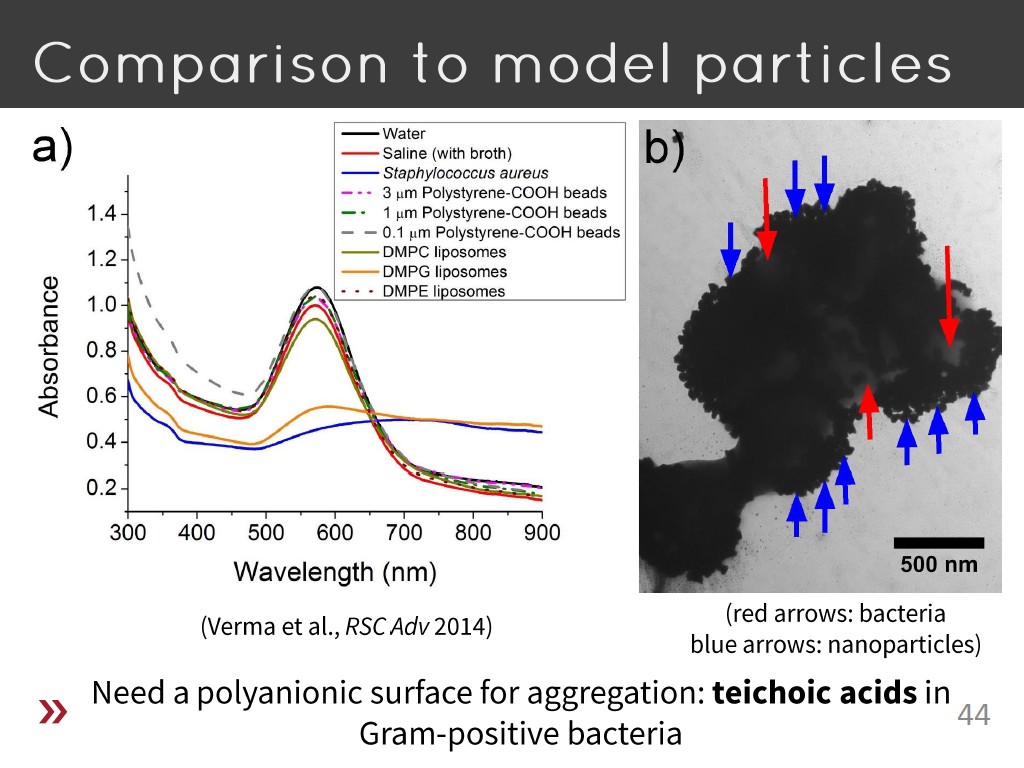 44. Comparison to model particles
1529.2292292292293
00:00/00:00
44. Comparison to model particles
1529.2292292292293
00:00/00:00 -
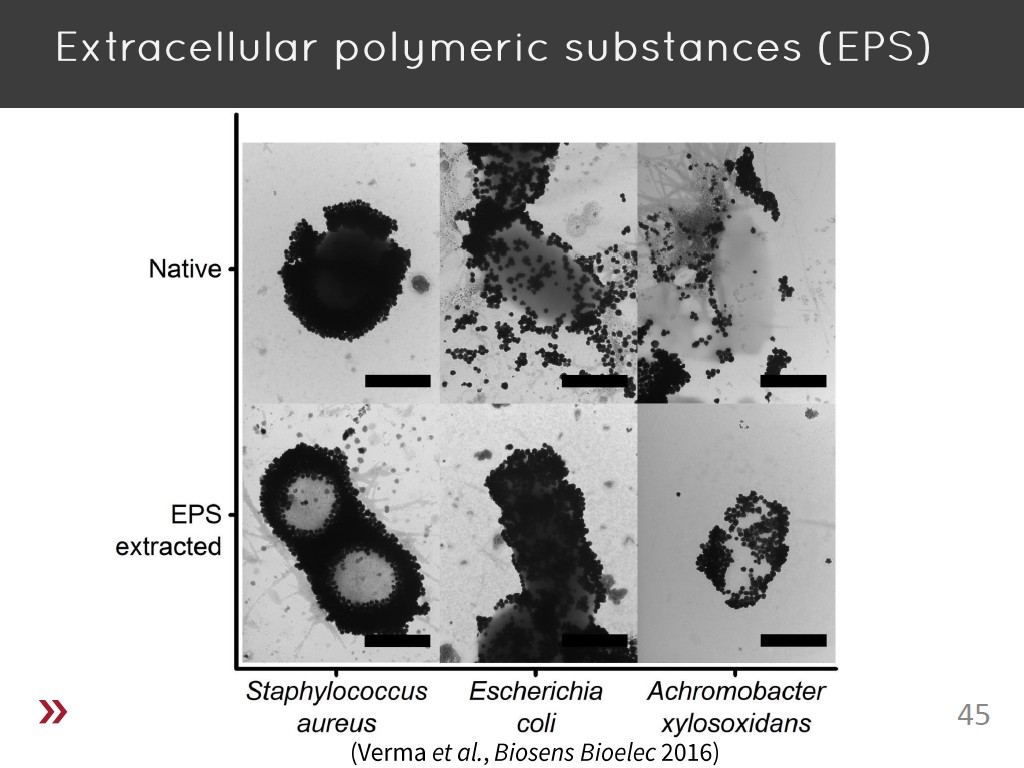 45. Extracellular polymeric substa…
1609.743076409743
00:00/00:00
45. Extracellular polymeric substa…
1609.743076409743
00:00/00:00 -
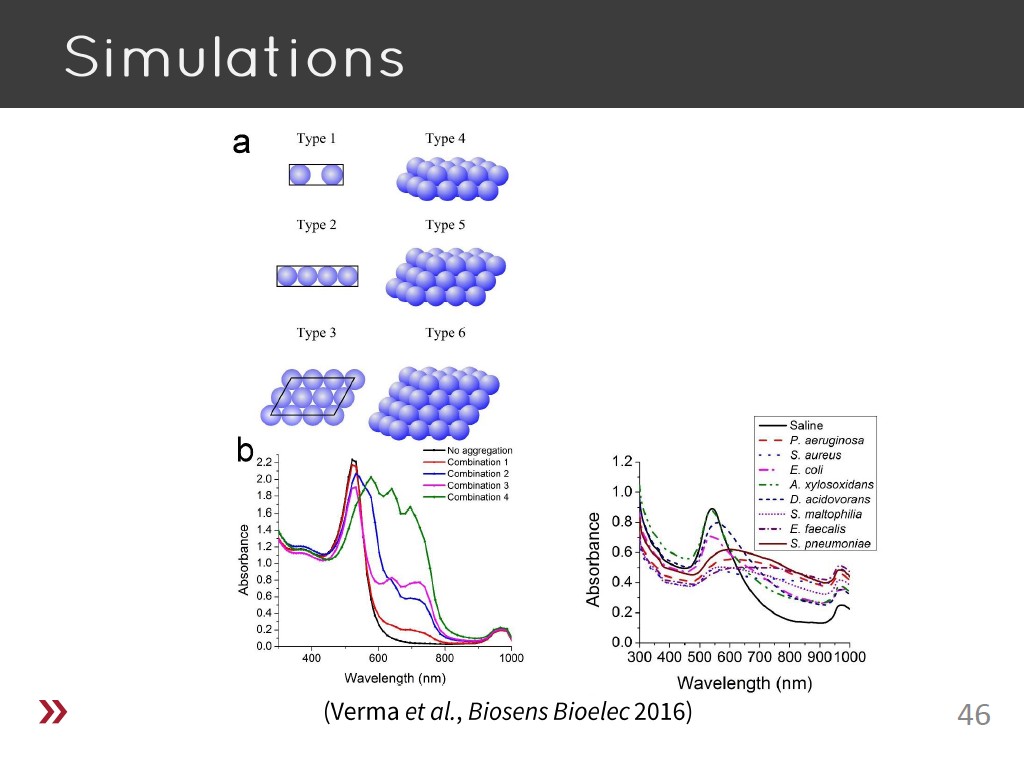 46. Simulations
1664.6646646646648
00:00/00:00
46. Simulations
1664.6646646646648
00:00/00:00 -
 47. Future Plans
1713.446780113447
00:00/00:00
47. Future Plans
1713.446780113447
00:00/00:00 -
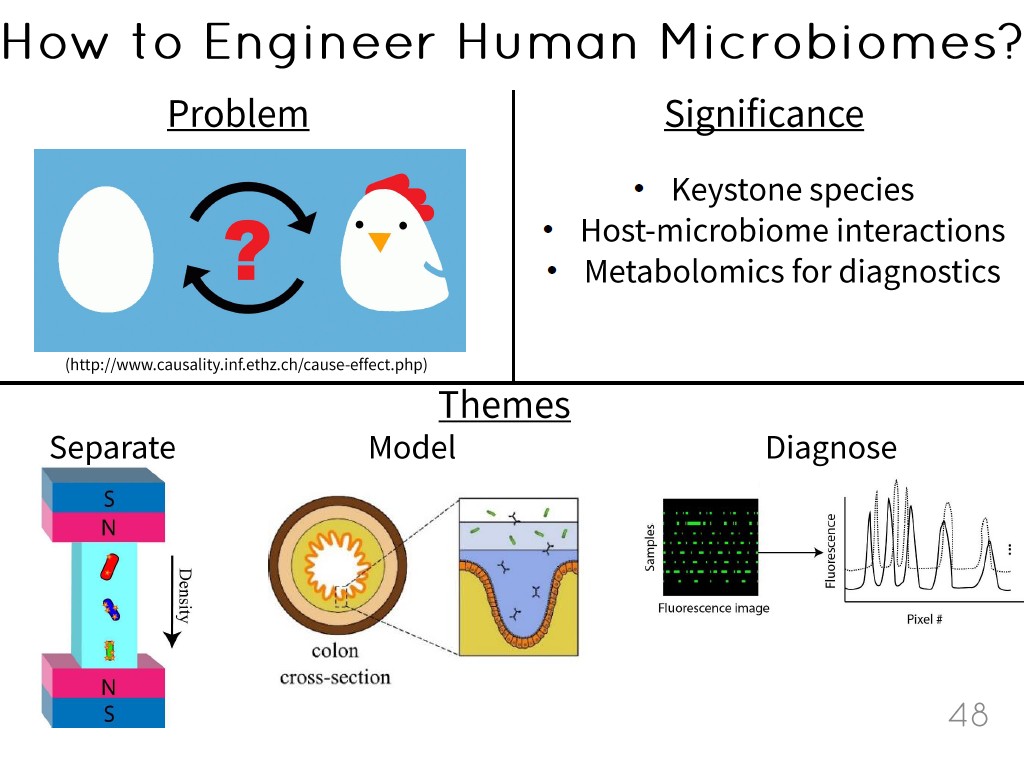 48. How to Engineer Human Microbio…
1720.2535869202536
00:00/00:00
48. How to Engineer Human Microbio…
1720.2535869202536
00:00/00:00 -
 49. Summary
1874.341007674341
00:00/00:00
49. Summary
1874.341007674341
00:00/00:00 -
 50. Acknowledgements
1906.0393727060396
00:00/00:00
50. Acknowledgements
1906.0393727060396
00:00/00:00 -
 51. Acknowledgements
1935.201868535202
00:00/00:00
51. Acknowledgements
1935.201868535202
00:00/00:00 -
 52. Thank You
1965.4988321654989
00:00/00:00
52. Thank You
1965.4988321654989
00:00/00:00
 Dr. Mohit Verma is a postdoctoral fellow in Professor George Whitesides' laboratory at Harvard University. Dr. Verma received his PhD in Chemical Engineering (Nanotechnology) in 2015 and his BASc in Nanotechnology Engineering in 2012 from the University of Waterloo (Canada). During his PhD, he conducted research on the use of gold nanoparticles for detecting bacteria and on the use of polymeric nanoparticles for delivering drugs, under the supervision of Professor Frank Gu. He received the Vanier Canada Graduate Scholarship from the Government of Canada and the Endeavour Research Fellowship from the Government of Australia (for spending four months at the University of New South Wales) during his PhD. He currently holds the Banting Postdoctoral Fellowship from the Government of Canada. His research interests include human microbiomes, biosensors, drug delivery, and soft robots.
Dr. Mohit Verma is a postdoctoral fellow in Professor George Whitesides' laboratory at Harvard University. Dr. Verma received his PhD in Chemical Engineering (Nanotechnology) in 2015 and his BASc in Nanotechnology Engineering in 2012 from the University of Waterloo (Canada). During his PhD, he conducted research on the use of gold nanoparticles for detecting bacteria and on the use of polymeric nanoparticles for delivering drugs, under the supervision of Professor Frank Gu. He received the Vanier Canada Graduate Scholarship from the Government of Canada and the Endeavour Research Fellowship from the Government of Australia (for spending four months at the University of New South Wales) during his PhD. He currently holds the Banting Postdoctoral Fellowship from the Government of Canada. His research interests include human microbiomes, biosensors, drug delivery, and soft robots.