You must login before you can run this tool.
Category
Published on
Abstract
Multi-species Biofilm on Wound Surface
Aneequa Sundus and Furkan Kurtoglu
Description
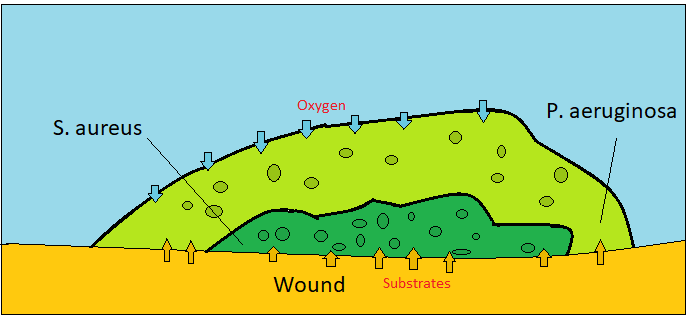
This tool aims to model two bacterial species which form a biofilm on the surface of a chronic wound as described in [4]. One species is Pseudomonas aeruginosa which is an aerobic species while the other one Staphylococcus Aureus is anaerobic. Aerobic species provides a cover from atmospheric oxygen which has a negative effect on anaerobic cells. Each agent utilizes glucose as a carbon source which is supplied at the bottom of the domain. This glucose are secreted by wound cells. In addition, air sources oxygen to the aerobic species.
The model is created to be used as a ENGR-599 class project. This model is developed by using agent-based modeling simulator PhysiCell [1]. The user interface is provided by xml2jupyter package [2] and uploaded to NanoHub nano-bio branch.
Chemical Compositions
In this model, there are three biochemical entities, which are glucose, oxygen, and ECM (extracellular matrix). We modeled a vertical environment where the bottom two layers of voxels have wound cells while resting of the upper surface in air. Glucose and oxygen diffuse voxels by an efficient chemical diffusion tool which is called BioFVM [3]. However, since the domain is not horizontal and BioFVM does not support different diffusion coefficients for different voxels, we are making corrections accordingly to not allow glucose diffusion to air.
The domain has two different voxel types which are Dirichlet Nodes and Non-Dirichlet Nodes. The Dirichlet nodes supply oxygen with a constant concentration. However, non-Dirichlet nodes enable to diffuse chemicals. There are two scenarios to be non-Dirichlet nodes. The first type of non-Dirichlet voxels is initialized at the bottom of the domain that has wound cells. But the other type is nodes that are converted from Dirichlet voxels. This conversion occurs by ECM level. If the concentration of ECM, which is secreted by bacterial species, is greater than 1 a.u, it became non-Dirichlet node which allows diffusion.
Oxygen:
This chemical is important for aerobic growth. Because it is sourced by air, Dirichlet nodes are important for oxygen. Dirichlet boundary condition is defined as 20 mmHg. Diffusion length is 100 micrometer.
Glucose:
Glucose is important for both species in the generic energy equation. Wound cells secrete glucose with 10 1/min rate and 10 a.u saturation density. Secreted glucose diffuses through non-Dirichlet nodes to source bacterial cells.
ECM:
ECM is the backbone of structure and a decision-maker biological entity for non-Dirichlet voxels, which has a very small diffusion length as 0.1 micrometers. ECM is secreted by bacterial cells.
Agent Cells
Bacterial cells are really small (1-micron diameter), so it poses difficulty in visualization when it is compared to wound cells. Furthermore, simulating bacterial cells with their actual size would result in a very large number of cells, hence increasing computational cost. Thus, our bacterial colonies are of the same size as that of wound cells for simplicity. They can be considered a collection of 100 bacterial cells in one species micro-colony. But it is assumed that one colony can include one type of bacteria.
Key features in agents
Birth & Death
Wound cells do not have any growth or death rule but bacterial cells can proliferate or die according to one generic energy equation. Each cell has cell data which is “energy”. This energy is created according to essential chemical concentrations. For an instant, the growth of P.aeruginosa is depended on glucose and oxygen while S.aureus uses only glucose. Energy can is used in constant rate for cellular events which depletes it. If the energy level is greater than the cycle energy threshold, bacterial cells go for the division. On the other hand, if the energy falls smaller than the death threshold, cells go for apoptosis. In addition to the use rate, there is another penalty term for anaerobic cells that relate to oxygen concentration. Generic Energy Rate which is applied for each cell is provided below.

E: energy (a.u)
t: time
b: energy creation rate (1/min)
alpha: energy creation rate modifier for aerobic species (dimensionless)
phi: energy creation rate modulator for aerobic species (dimensionless)
beta: energy creation rate modifier for anaerobic species (dimensionless)
chi: energy creation rate modulator for anaerobic species (dimensionless)
gamma: energy usage rate modulator for both species (dimensionless)
d: energy usage rate (1/min)
rho: energy penalty for anaerobic species (dimensionless)
Both species are non-motile as we assume that they are big colonies and they do not cooperatively move to somewhere. In addition, wound cells are non-movable which not allowing bacterial cells pushing the wound cells.
Agent types
Wound Cells(black cells)
These are non-movable cells present on the bottom surface through simulation. They are acting as glucose source. They are colored black in simulation
Staphylococcus Aureus (Anaerobe)(Red Cell)
Bacterial cells that can only survive if they are surrounded by Aerobic cells.
Pseudomonas Aeruginosa (Aerobe)(Blue Cells)
These cells secrete ECM in environment.They uptake glucose and oxygen from the environment
Tool Instructions
-
Modify the core parameters in the tool, such as, domain size, saving options, simulation time.
-
Change the model parameters accordingly.
-
For the seeding method: You can use 4 options for seeding
-
vertical: Seeding species side by side
-
horizontal: Seeding species on top of them
-
random: Randomly seeding cells
-
box: Seeding aerobe that covers
-
-
References
[1] Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://doi.org/10.1371/journal.pcbi.1005991
[2] Heiland R, Mishler D, Zhang T, Bower E, Macklin P (2019, in preparation) Xml2jupyter: Mapping parameters between XML and Jupyter widgets. J Open Source Software.
[3] Ghaffarizadeh A, Friedman SH, Macklin P (2016) BioFVM: an efficient, parallelized diffusive transport solver for 3-D biological simulations. Bioinformatics 32(8):1256-8. https://doi.org/10.1093/bioinformatics/btv730
Cite this work
Researchers should cite this work as follows: