You must login before you can run this tool.
Category
Published on
Abstract
Introduction
Synthetic biology and other bioengineering techniques are increasingly creating novel compounds that can target and tune specific aspects of cell phenotype (behavior), such as proliferation, adhesion, motility, and metabolism. This presents both key opportunities and challenges: What if we could tune what a therapeutic does, and where it is delivered? Which therapies would be worth delivering? What are the key design parameters?
The aim of this tool is to explore these what if questions: users can set nanoparticle, tissue, cell, and drug response parameters for a growing tumor, and see for themselves how things could play out for their design choices.
Modeling description
In this model, We simulate a tumor growing in the microenvironment. We first follow experimental data to fit the dynamics of celluar update or internalization. We also propose a dynamics equation for drug release to transit NPs while release drug in of individual cells, and then explore the tumor response to drug. Our model introduces a novel tracking of nanoparticle populations in each individual cells, allowing better modeling of drug release by internalized nanoparticles and long-term therapeutic implications.
Modeling approach
We used agent-based models to simulate the tumor cells growing in the microenvironment. The modeling mainly involves two processes: pharmacokinetics and pharmacodynamics.
Nanoparticles and pharmacokinetics:
In the microenvironment, NPs are released in dose (by setting the initial condition and Dirichlet boundary condition). Tumor cells uptake the NPs with the following dynamics equation. In the uptake equation, the celluar uptake rate Ui is proportional to internalization rate rI, and inverse with the number of internalized nanoparticles nI, with a capacity of n*. Here, we also introduce a drug release model, in which NPs go through several transition processes, with releasing drug in the process, where the transition dynamics for NPs in jth: nj is related with transit from last process rj-1nj-1 and next process rjnj, and transit numbers N and the intial drug on one nanoparticle C*; for the release rate rj is proportional to the remained drug in the NPs within jth transition process Cj and inverse with the drug concentration c inside of each cells, see the following schematic:
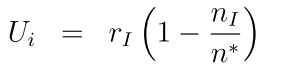
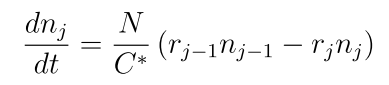

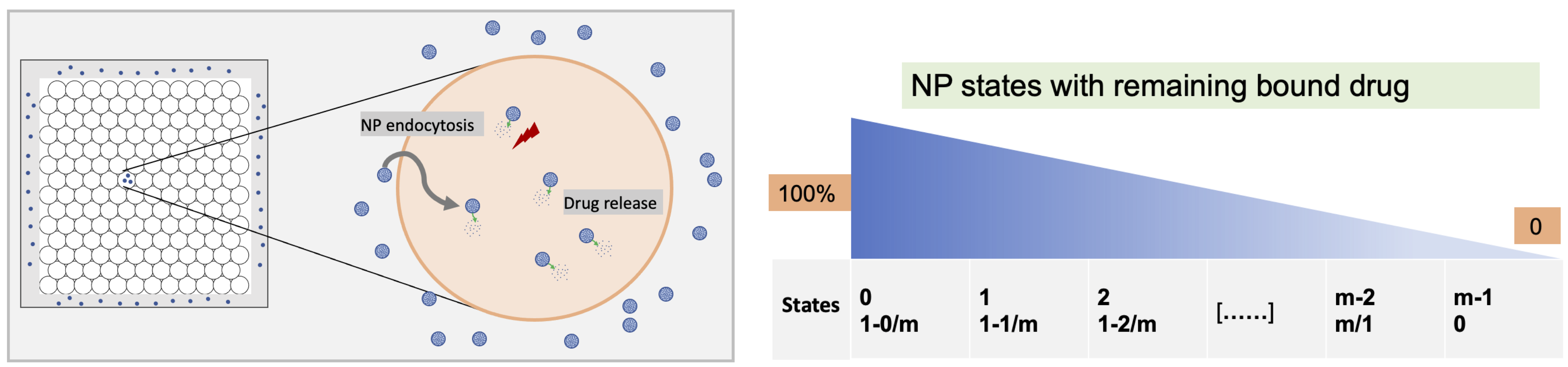
Overview of computational diagram and NP states with bound drug
Cells and drug pharmacodynamics:
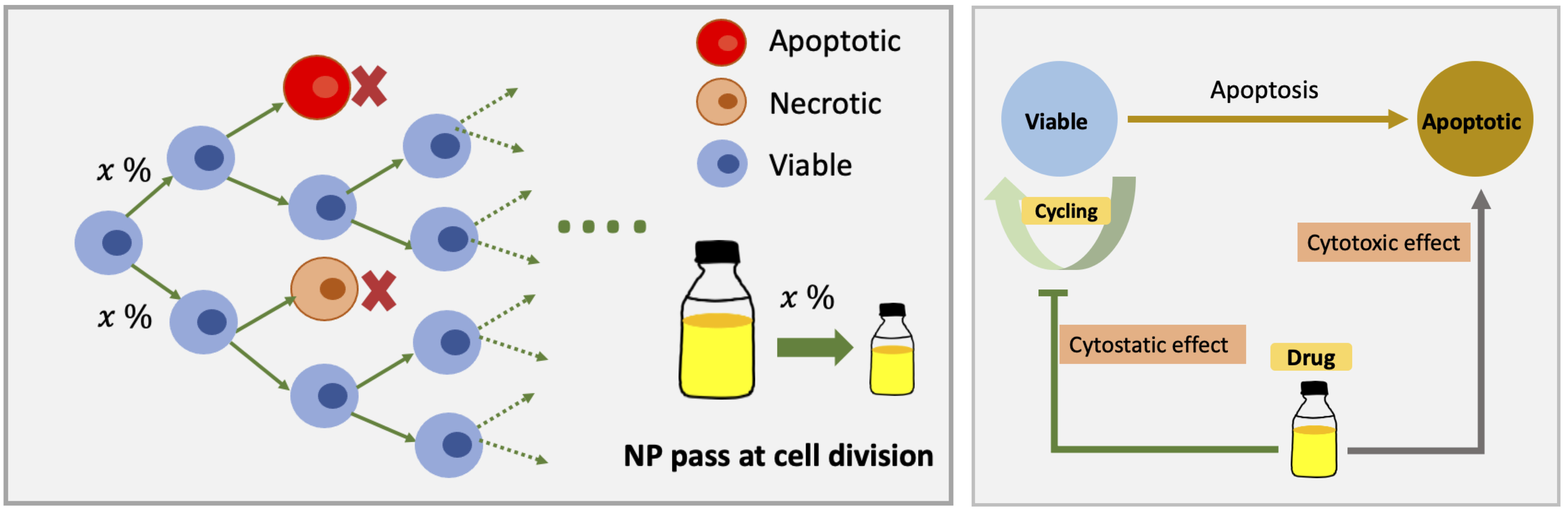
NP "inherited" at cell division and Drug effect on tumor cell phenotype
Without drug:
Cells will be assigned a wild-type microenvironment-dependent phenotype P0(E), where P is the vector of phenotype properties, and E is the cell's local microenvironmental conditions. As in PhysiCell [1], cell cycle entry increases with oxygenation, apoptosis occurs at a steady background rate throughout viable tissue, and necrosis increases with oxygen depletion.
With drug:
We model the maximum phenotype response vector PM as tumor response to drug. We assume drug has a Hill coefficient (h) and half-max effect drug concentration (c0.5). We use a Hill effects model:
E = ch / ( c0.5h + ch).
Note that the code will also allow us to replace drug with an AUC (area-under-the-curve) model ∫c(s) ds (from 0 to t). Also, the code will allow an active damage and repair model, based upon the drug concentration.
For each cell's current effect E, we set its phenotype to:
P = (1-E) P0(E) + E PM
Example treatment results:
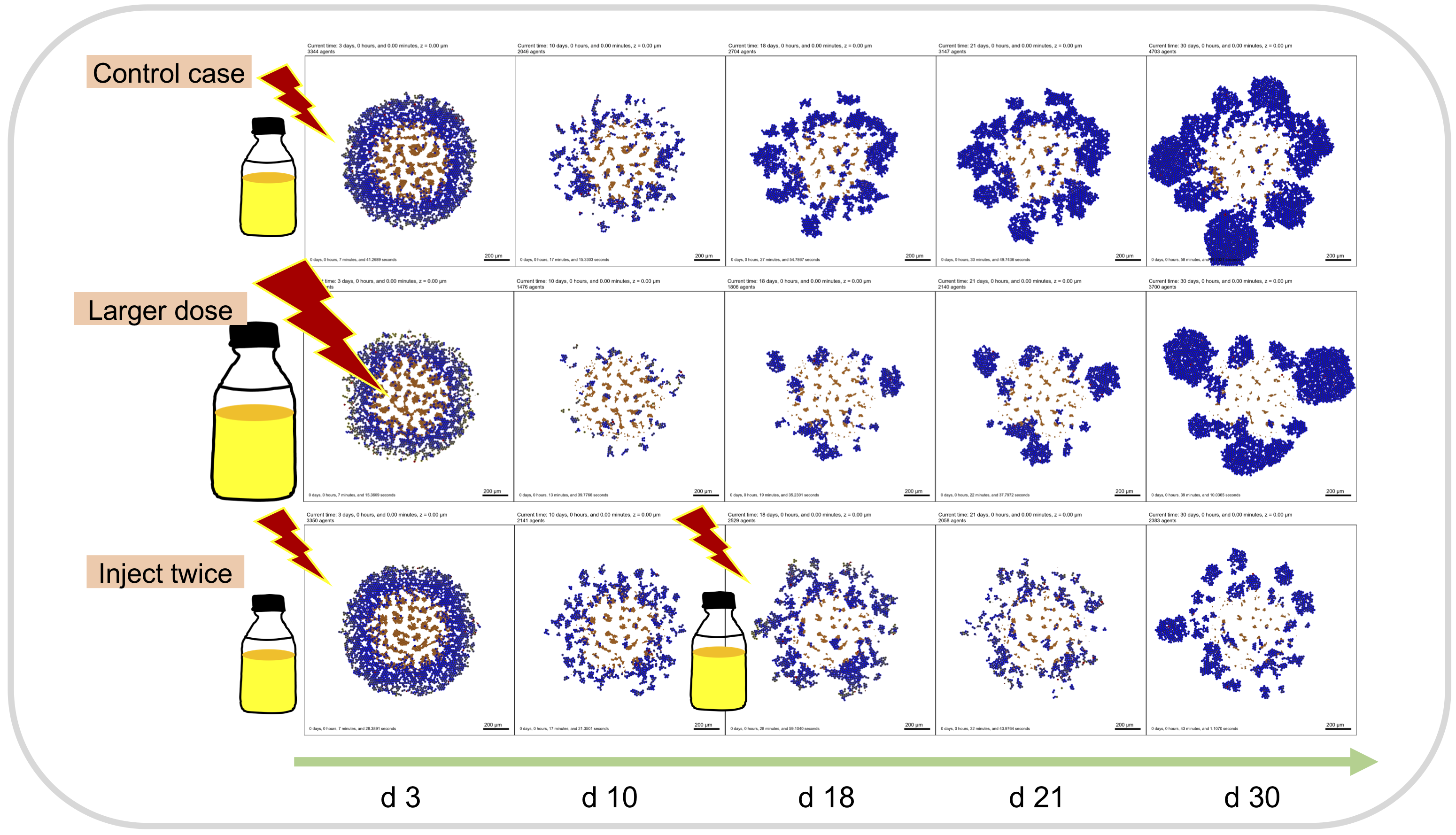
Chemical components:
All chemical signals move by chemical diffusion in the simulated environment, using BioFVM [2] to solve the reaction-diffusion equations. A key property is the diffusion length scale. Diffusion (with a diffusion coefficient D) helps spread a signal over large distances, while decay (and uptake) (with coefficient λ) eliminates the signal to slow its spread. These effects compete to determine the characteristic distance Lthat a chemical signal travels.
In our model, oxygen and NPs are diffused with BioFVM [2] by setting the initial condition and Dirichlet boundary condition.
Cell agents:
Key features in the agent-based models:
Birth and death:
Cells can proliferate (divide into two daughter cells of half size), with division rates regulated by cell rules. In our model, cells continuously work to grow towards a "target" mature volume.
Note that birth and death are stochastic events for each cell agent: if such a process occurs at rate r, then between now (t) and soon (t + Δt), the probability of the event occurring for that agent is rΔt.
In our model, tumor cells would experience normal apoptosis at a steady background rate throughout viable tissue, and necrosis increasing with oxygen depletion in no drug microenvironment. With drug, the phenotype of tumor cells changes with interpolation with background rate and max effect as described in section of Modeling approach.
Secretion and uptake:
Cells can secrete chemical factors, or they can remove them (i.e., consume or uptake). This can contribute to gradients in chemical factors, and it's a key method of communication between cells. Moreover, cells can "sample" the chemical state of their surrounding environment. Secreting is sending a signal. Uptaking and sampling is receiving a signal.
In our model, tumor cells uptake the NPs, and then NPs release drug inside of each cells, see details in section of Modeling approach.
Biased migration:
Motile cells perform biased migration. After traveling for Tpersistence time (the mean persistence time), they choose a new migration direction dmigrate. Based on the environmental conditions, the cell chooses a directional bias (intended direction) dbias and a random unit vector r. The motile direction dmigrate is then determined by according to:
dmigrate = b dbias + (1-b) r.
Here, b is a bias parameter (between 0 and 1) that determines how strongly biased migration is towards dbias. If b =1, then migration is deterministic towards dbias. If b = 0, then migration is completely random (Brownian).
Once the migration direction is chosen, it is normalized, and multiplied by the cell's migration speed.
In out model, we simplified cells as immotile.
Mechanics:
Cell agents can stick to one another within a prescribed interaction distance (some multiple of their radius), and they can exert a pushing force on neighbors. PhysiCell [1] uses potential functions to implement these simple mechanics. Notably, PhysiCell is an off-lattice model, meaning that cells can have variable sizes, and can move freely without grid artifacts.
They aren't required to move some prescribed number of spatial steps. They also can divide without checking for an open neighbor "site". Instead, they can divide and push their neighbors out of the way.
Cell-cell interactions:
In the model, we currently only have one type of cells (tumor cells).
Tumor cells (blue)
The built-in "live" cell cycle model is used in our model, with the default models of apoptosis and necrosis. Thus, each cell agent can be Live, Apoptotic, or Necrotic. As explained in the Cells and drug pharmacodynamics section, the phenotype of tumor cells would be changed based on the drug concentration with a Hill effects model.
Basic instructions
Modify parameters in the "Config Basics", "Microenvironment", or "User Params" tabs. Click the "Run" button once you are ready.
To view the output results, click the "Out: Plots" tab, and move the slider bar to advance through simulation frames. Note that as the simulation runs, the "# cell frames" field will increase, so you can view more simulation frames.
If there are multiple substrates defined in the Microenvironment, you can select a different one from the drop-down widget in the Plots tab. You can also fix the colormap range of values.
Note that you can download full simulation data for further exploration in your tools of choice. And you can also generate an animation of the cells to play in the browser and, optionally, download as a video.
About the software
This model and cloud-hosted demo are part of the education and outreach for the IU Engineered nanoBIO Node and the NCI-funded cancer systems biology grant U01CA232137. The models are built using PhysiCell: a C++ framework for multicellular systems biology [1] for the core simulation engine and xml2jupyter [3] to create the graphical user interface (GUI).
Recent changes
0.9.1-0.9.2
- Add divider description for parameters in User Params of GUI
- Add Animate tap to generate video in GUI
0.9
- Updated the xml2jupyter
- Updated the PhysiCell to 1.6.0 version
- Modified model as one type of NPs in the microenvironment
- Proposed a drug release model
- Tracked internalization NPs and drug concentration in each cells
0.8.1
- Fixed a bug in the simulation code that sometimes caused a segfault when running with motile cells on multiple threads.
- Enabled cell motility, plus related parameters, in the GUI.
- Provided functionality to submit batch jobs and visualize their results.
- Provided cached results for both batch and interactive runs.
- Provided a "Load Config" dropdown widget to access cached results.
- Provided download buttons for (compressed) files associated with plots.
- Rearranged widgets in the Substrate Plots tab to more closely match Cell Plots tab.
- Improved the tool startup process, trying to avoid a blank screen.
0.7.0
- Allow selecting previously computed output (directories)
- Save/remember min/max for substrate ranges.
0.6.3
- Added plot titles to substrate plots.
- Added additional colormap palettes.
- Using checkboxes in plots updates displayed plots.
0.6.2
- Updated substrate plots to allow a fixed min/max range.
- Improved cell visualizations with (optional) black outlines to improve contrast with white background.
- Improved cell glyph (marker) size calculation.
0.6.1
- Disabled chemotaxis due to compiler issues.
- Minor usability refinements.
0.6.0
- Usability refinements.
0.5.0
- Initial public release.
Future improvements
User interface improvements:
- Enable motility (after migration to platform with gcc 6.0 or greater)
- Better defaults on slider bars for substrate visualization
- Further refinements based on user feedback
Scientific improvements:
- Integrate/transfer techniques applied in viral growth inhibition [4] to this model
- Track NPs and drug from parents cells to daughter cells when divide
- Include NP binding/unbinding to ECM
- Work collaboration with experimental peers to improve the model parameters
- Optimize model parameters and probably add deep learning method (e.g. LSTM) to predict the tumor growth within long term period
Powered by
This software is powered by PhysiCell [1-2], a powerful simulation tool that combines multi-substrate diffusive transport and off-lattice cell models. PhysiCell is BSD-licensed, and available at:
- GitHub releases: https://github.com/MathCancer/PhysiCell/releases
- SourceForge downloads: https://sourceforge.net/projects/physicell/
It is a C++, cross-platform code with minimal software dependencies. It has been tested and deployed in Linux, BSD, OSX, Windows, and other environments, using the standard g++ compiler.
See http://PhysiCell.MathCancer.org.
The Jupyter-based GUI was auto-generated by xml2jupyter [3], a technique to create graphical user interfaces for command-line scientific applications.
Bio
To learn more about our work, please visit MathCancer.org.
Credits
Scientific Staff
Yafei Wang, PhD candidate, Intelligent Systems Engineering, Indiana University.
- Develop the nanotherapy model
Randy Heiland, Research Associate, Intelligent Systems Engineering, Indiana University.
- Lead software architect
Paul Macklin, Ph.D. , Associate Professor, Intelligent Systems Engineering, Indiana University.
- Scientific lead, developed PhysiCell simulation model
The following undergraduate students have contributed to this project:
Jupyter notebook and UI development
- Eric Bower (IU Intelligent Systems Engineering, B.S., 2018-present)
- Daniel Mishler (IU Intelligent Systems Engineering, B.S., 2018-present)
- Tyler Zhang (IU Intelligent Systems Engineering, B.S., 2018-present)
Sponsored by
NSF EEC-1720625. Network for Computational Nanotechnology - Engineered nanoBIO Node
References
[1] Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://doi.org/10.1371/journal.pcbi.1005991
[2] Ghaffarizadeh A, Friedman SH, Macklin P (2016) BioFVM: an efficient, parallelized diffusive transport solver for 3-D biological simulations. Bioinformatics 32(8):1256-8. https://doi.org/10.1093/bioinformatics/btv730
[3] Randy Heiland, Daniel Mishler, Tyler Zhang, Eric Bower, Paul Macklin (2019) xml2jupyter: Mapping parameters between XML and Jupyter widgets. https://joss.theoj.org/papers/10.21105/joss.01408
[4] M Getz et al (2020) Iterative community-driven development of a SARS-CoV-2 tissue simulator. bioRxiv preprint 2020.04.02.019075. https://doi.org/10.1101/2020.04.02.019075
[5] Wang Y (2022). Multicellular simulation of heterogeneous tissues with multi-scale system effects: A high-throughput agent-based modeling approach. ProQuest Dissertations and Theses, [Order No. 29252377]. Indiana University
Publications
No publications have cited this tool. Yet. :-)
Cite this work
Researchers should cite this work as follows:
-
This tool is still in preparation for submission. For now, please cite:
Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://doi.org/10.1371/journal.pcbi.1005991